Abstract
In this study, the complete mitochondrial (mt) genome of Chlamydotis macqueenii was sequenced through the Illumina sequencing method, and the mitochondrial genome map was constructed after de novo assembly and annotation. This intact mt genome (16,821 bp) has a typical gene repertoire in vertebrates, with 13 protein-coding genes (PCGs), 22 tRNAs, and 2 rRNAs. The overall GC content of this mitogenome is 44.5%. By phylogenetic analysis using IQTREE, C. macqueenii showed the closest relationship with Otis tarda in the subclade of Otididae.
In this study, we assembled and analyzed the mitochondrial genome of C. macqueenii based on the next-generation sequencing method.
Tissue samples of C. macqueenii sequenced in this study were obtained from the dead individual occasionally found in Gobi Desert located in Gansu province, China (39°57'N, 105°72'E). Genomic DNA was extracted from tissues fixed in ethanol using QLAGEN DNEasy Extraction Kit, following the manufactures instructions. The isolated DNA was stored in the sequencing company (HuiTong Tech, Shenzhen, China). Purified DNA was fragmented and used to construct the sequencing libraries following the instructions of NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). Sequencing was done on an Illumina HiSeq2500 platform (Illumina, San Diego, CA). The clean reads were assembled by SPAdes 3.11.1 (Bankevich et al. Citation2012) with default settings.
We used other mitochondrial genomes from Neognathae as seeds to collect the mitochondrial contigs as much as possible. To fill the gap, Price (Ruby et al. Citation2013) and MITObim v1.8 (Hahn et al. Citation2013) were applied and Bandage (Wick et al. Citation2015) was used to construct the circular assembly path guided by these positions of long scaffolds. The complete sequence was primarily annotated by ORF prediction in Unipro UGENE (Okonechnikov et al. Citation2012) combined with manual correction. All tRNAs were confirmed using the tRNAscan-SE search server (Lowe and Eddy Citation1997). This complete mitochondrial genome sequence was submitted to GenBank under the accession numbers of MK714019.
The mitochondrial genome of C. macqueenii is a typical circular structure of 16,821 bp with a GC content of 44.5%. This mitochondrial genome contains a total of 37 genes, including 13 PCGs, 2 rRNA genes (12S and 16S ribosomal RNA), and 22 tRNA genes.
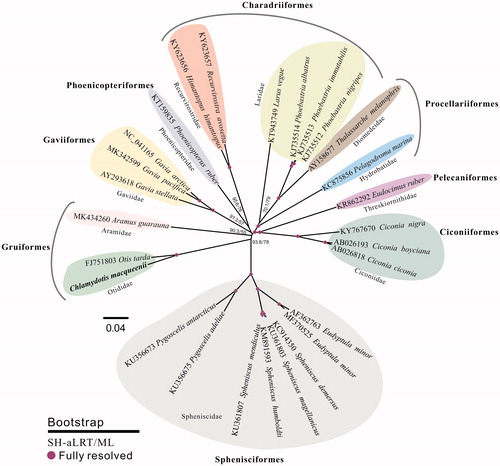
For phylogenetic analysis, another 26 published complete mitochondrial sequences of Neognathae were retrieved from the NCBI database to build genome-wide alignment by HomBlocks (Bi et al. Citation2018), with default parameters. Then, the resulting alignments with 14,912 positions were imported into IQ-TREE version 1.6.6 (Nguyen et al. Citation2015), according to the optimal substitution models under the rapid bootstrap algorithm (1000 replicates), to refer phylogenetic relationships among 27 Neognathae mt genomes. To test support for the branch points of each gene trees, nonparametric branch support tests based on the Shimodaira–Hasegawa-like approximate likelihood ratio test (SH-like aLRT) procedure were also performed in the same run. As shown in , the phylogenetic positions of these 27 mt genomes were successfully resolved with full bootstrap supports across almost all nodes. Chlamydotis macqueenii, belonging to the Otididae, exhibited the closest relationship with Otis tarda in the subclade of Otididae.
Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Computational Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Okonechnikov K, Golosova O, Fursov M, the UGENE team. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 28:1166–1167.
- Ruby JG, Priya B, Joseph LD. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3: Genes, Genomes, Genetics. 3:865–880.
- Wick RR, Schultz MB, Schultz J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.