Abstract
The complete mitogenome of Cyclosa japonica (GenBank accession number MK512575) is 14,687 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes, and a putative control region. Its gene content and organization are identical with other spider mitogenomes. The overall base composition was as follows: A (35.61%), T (37.35%), C (10.87%), and G (16.18%), with an A + T bias (72.96%). Seven tRNAs (trnM, trnW, trnK, trnE, trnF, trnH, and trnT) lacked the TΨC arm stem, while two tRNAs (trnS1 and trnS2) lacked the dihydrouracil (DHU) arm. Phylogenetic tree based on concatenated amino acid sequences of 13 PCGs shows that C. japonica is closely related to Cyclosa argenteoalba, which accord with morphological classification.
Keywords:
The orb-weaving spider, Cyclosa japonica (Araneae: Araneidae), is an important predator of several economical agricultural pests (Wang et al. Citation1996; Platnick Citation2015). This species is mainly distributed in China, Russia, Korea and Japan. These spiders weave small webs in paddy fields, trees, and tea bushes. In this study, adult individuals of C. japonica were collected from Maolan Nature Reserve in Libo country, Guizhou Province, China (E107°58′, N25°15′), and preserved in the spider specimen room of Guiyang University with an accession number GYU-GZML-14.
The complete mitogenome sequence of C. japonica is 14,687 bp in length and is deposited in GenBank under Accession number MK512575. It harbors 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rrnL and rrnS), and a putative control region. The gene content and organization of C. japonica are identical with other spider mitogenomes (Li et al. Citation2016; Xu et al. Citation2019). Twenty-two genes were encoded on the major strand (J-strand), while others were transcribed on the minor strand (N-strand). The overall base composition was as follows: A (35.61%), T (37.35%), C (10.87%), and G (16.18%), with an A + T bias (72.96%). This mitogenome presented a negative AT-skew (–0.024) and a positive GC-skew (0.196).
The C. japonica mitogenome displays gene overlap in 28 bp in seven locations. The longest overlap is 16 bp in length and located between cob and trns2. The intergenic spacer sequences were spread over 24 regions ranging in size from 1 to 39 bp. The 22 tRNAs had a total of 1297 bp, and their individual lengths ranged from 50 bp (trnC) to 67 bp (trnL2 and trnH). Nine tRNAs appear to be truncated and lack the potential to fold into the cloverleaf secondary structure. Seven of them (trnM, trnW, trnK, trnE, trnF, trnH, and trnT) lacked the TΨC arm stem, and two tRNAs (trnS1 and trnS2) lacked the dihydrouracil (DHU) arm.
The rrnL and rrnS were located between trnL1 and trnQ and separated by the trnV, with a length of 1017 and 684 bp, respectively. The control region was located between trnQ and trnM with a length of 1145 bp, and the A + T content was 69.26%. Three PCGs (cox3, nad6, and nad1) terminate with TAG, two PCGs (nad4 and nad4L) use T and the rest of the genes stop with TAA. Nine PCGs start with typical ATN start codons (ATA, ATT, and ATG), three genes (cox2, cox3, and nad6) use TTG and cox1 is initiated with TTA.
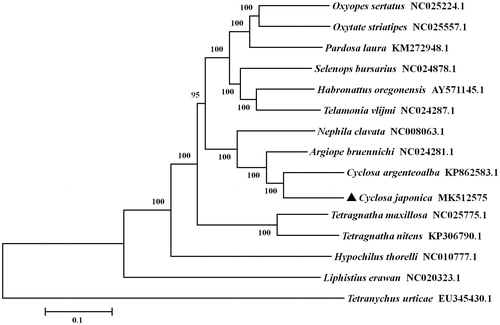
To validated with the phylogenetic position of C. japonica, a neighbor-joining phylogenetic tree was constructed using the concatenated amino acid sequences of 13 PCGs with MEGA6 (Tamura et al. Citation2013). The result showed that C. japonica is closely related to Cyclosa argenteoalba (), which accord well with traditional morphological classification.
Figure 1. Phylogenetic tree showing the relationship between Cyclosa japonica and 13 other spiders based on neighbor-joining method. Spider determined in this study is labeled with triangle symbol. GenBank accession numbers of each species are listed in the tree. Tetrancychus urticae was used as an outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Li C, Wang ZL, Fang WY, Yu XP. 2016. The complete mitochondrial genomes of two orb-weaving spider Cyrtarachne nagasakiensis (Strand, 1918) and Hypsosinga pygmaea (Sundevall, 1831) (Araneae: Araneidae). Mitochondrial DNA Part A. 27:2811–2812.
- Platnick NI. 2015. The world spider catalog, version 13.5. American Museum of Natural History. http://research.amnh.org/iz/spiders/catalog/.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang HQ, Yan HM, Yang HM. 1996. Studies on the ecology of spiders in paddy fields and utilization of spiders for biological control in China. Sci Agr Sin. 29:68–75.
- Xu KK, Yang WJ, Yang DX, Li C. 2019. The complete mitochondrial genome sequence of Neoscona multiplicans (Chamberlin, 1924) (Araneae: Araneidae). Mitochondrial DNA Part B. 4:201–202.
