Abstract
Capsella rubella (C. rubella) is a highly selfing plant species, mostly occurring in southern and western Europe. As a member of the closest well-characterized genus (Capsella) to Arabidopsis thaliana (A. thaliana), C. rubella has been studied by botanists in recent years. In this study, we successfully assembled the complete mitochondrial (mt) DNA sequence of C. rubella into a circular genome of 287,405-bp length, encoding 31 protein-coding genes (PCGs), 21 tRNA genes, and 3 rRNA genes. The GC content of the C. rubella mt genome is 44.74%, which shows a prevalent GC content within Brassicaceae plant species. The reconstructed neighbour-joining tree supported that C. rubella was evolutionarily close to A. thaliana and Brassica napus (B. napus), both of which belong to Brassicaceae. This worthwhile research of C. rubella mt genome will not only provide a valuable reference for model plant A. thaliana, but also contribute to the study of the evolution of self-incompatibility to self-compatibility.
Capsella rubella belongs to the genus Capsella, which is the closest well-characterized genus to A. thaliana. As we all known, A. thaliana is a popular model organism in plant biology and genetics, so the research of C. rubella will provide valuable information for the plant species. Additionally, C. rubella is always considered as a model plant to investigate the evolution of self-incompatibility into self-compatibility in plant reproduction (Ya-Long et al. Citation2009). Capsella rubella, the highly selfing plant species, is found mostly in southern and western Europe. We have found the complete nuclear and chloroplast genomes of C. rubella to be assembled; however, the mt genome has not yet been assembled (Slotte et al. Citation2013; Wu Citation2015). The main function of mitochondria is to convert energy from biomass into chemical energy in living cells, and play significant roles in plant productivity, development and phylogeny (Ye et al. Citation2017). The structures and sizes of mitochondria are highly dynamic (Richardson et al. Citation2013; Bi et al. Citation2016), even in the same family, so that there are only 245 plant-assembled mt genomes deposited in GenBank Organelle Genome Resources.
The C. rubella sample used for extraction and sequencing in our study was collected in Monte Gargano, Italy (Geographic coordinate: 41°44′02.1″N 15°45′00.0″E). Genomic DNA was extracted from the collected C. rubella sample according to the method described by Slotte et al. (Citation2013), and then deposited at the DoE JGI in Walnut Creek, California. In our study, we successfully assembled the complete mt DNA sequence of C. rubella into a 287,405-bp length circular genome according to the method described by Wang et al. (Citation2018). After that, we used the online program MITOFY to annotate the assembled mt genome (Alverson et al. Citation2010), and then submitted the annotated genome to GenBank with the accession number MH624151.
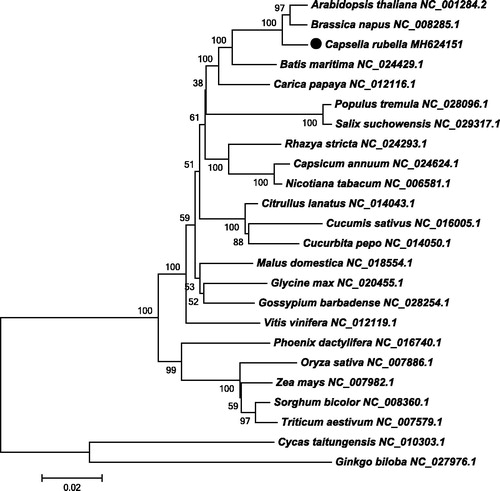
The structures and gene orders of plant mitochondria are extremely dynamic even in the same family, but the GC contents are conserved in the same order or family. The overall base composition of the C. rubella genome is as follows: A: 27.73%, C: 22.34%, G: 22.40%, T: 27.53%, and the GC content is 44.74%, which is a prevalent value in the mustard family, Brassicaceae. A total of 55 genes were annotated in the C. rubella mt genome, including 31 PCGs, 21 tRNA genes, and 3 rRNA genes. Codon analyses showed that ATG was the most used start codon in PCGs, except the NADH dehydrogenase nad1 use ACG (C to U RNA-editing) as its start codon, and the maturases matR and transport membrane protein mttB cannot determine its start codon. In the same manner, we also found three different types of stop codons in the mt PCGs, including TAG (5 genes: atp1, matR, mttB, nad7 and rps3), TGA (8 genes: atp8, atp9, ccmFn, cob, cox3, nad4, rpl2 and rps12), and TAA (18 genes: atp4, atp6, ccmB, ccmC, ccmFc, cox1, cox2, nad1, nad2, nad3, nad4L, nad5, nad6, nad9, rpl5, rpl16, rps4 and rps7). Additionally, we also identified 32 exons and 18 introns in nine PCGs (ccmFc, cox2, nad1, nad2, nad4, nad5, nad7, rpl2 and rps3). To obtain an accurate phylogenetic position of C. rubella, we reconstructed a neighbor-joining tree based on 23 conserved mt PCGs of 24 plant species in MEGA6 (Tamura et al. Citation2013). As shown in , the neighbor-joining tree illustrated that C. rubella was evolutionarily close to the clade composed of Arabidopsis thaliana and Brassica napus, and they all belong to the mustard family, Brassicaceae.
Acknowledgements
We would like to acknowledge the DoE JGI in Walnut Creek for their assistance in sampling the material of Capsella rubella.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27:1436.
- Bi C, Paterson AH, Wang X, Xu Y, Wu D, Qu Y, Jiang A, Ye Q, Ye N. 2016. Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches. BioMed Res Int. 2016:5040598.
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11:29.
- Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL, Steige K, Platts AE, Escobar JS, Newman LK, et al. 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 45:831–835.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang X, Cheng F, Rohlsen D, Bi C, Wang C, Xu Y, Wei S, Ye Q, Yin T, Ye N. 2018. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic Res. 5:3.
- Wu Z. 2015. The complete chloroplast genome of Capsella rubella. Mitochondrial DNA. 27:1.
- Ya-Long G, Bechsgaard JS, Tanja S, Barbara N, Martin L, Detlef W, Schierup MH. 2009. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci USA. 106:5246–5251.
- Ye N, Wang X, Li J, Bi C, Xu Y, Wu D, Ye Q. 2017. Assembly and comparative analysis of complete mitochondrial genome sequence of an economic plant Salix suchowensis. PeerJ. 5:e3148.