Abstract
Angelica sinensis is one of the most widely used Chinese medicinal herb. It has been cultivated for over 900 years, and the wild resource has been on the verge of extinction owing to over-exploitation. In the present study, we assembled and characterized the complete chloroplast (cp) genomes of two wild and one cultivated individuals of A. sinensis for future genetic studies. These plastome genomes varied from 142,484 to 142,529 bp in size, and overall GC contents were about 37.5%. Phylogenetic analysis and comparison on the cp genome structure reveal that the wild A. sinensis sampled should not be the wild ancestor, but the escape of the cultivated.
Angelica sinensis (Oliv.) Diels is a famous traditional Chinese medicinal herb and its dried root has been used in China for more than 2000 years. Although it has been cultivated on a large scale since the Song Dynasty (over 900 years ago), the wild A. sinensis has been on the verge of extinction owing to over-exploitation and only occasionally found in a few areas of Sichuan, Gansu, Shaanxi and Tibet Provinces (Sun et al. Citation2009; Zhang et al. 2009). Wild populations of a crop are the natural gene pool for breed improvement of the crop, and genetic studies on wild populations will give insights for germplasm innovation of cultivated plants (Zhang et al. Citation2017). Therefore, we assembled and compared complete chloroplast genomes of the wild and cultivated A. sinensis.
We collected two wild individuals of A. sinensis from Maye Village, Minxian County, Ganshu Province (103°50′52.76″E, 34°23′11.78″N) (A. sinensis_WMX) and Emei Mountain, Emeishan City, Sichuan Province (103°23′12.86″E, 29°33′18.21″N) (A. sinensis_WEM), respectively. One cultivated individual was obtained from Machang Village, Heqing County, Yunnan Province (100°04′6.49″E, 26°28′32.04″N) (A. sinensis_CHQ). All vouchers are deposited in the museum of Yunnan University of Chinese Medicine. Total genomic DNA was isolated with a modified CTAB method (Doyle and Doyle Citation1987). The chloroplast genome was amplified using nine universal primer pairs (Yang et al. Citation2014), and sequencing was performed on the Illumina Hiseq 2000 platform. The plastome genome was assembled using Geneious 8.1 (Kearse et al. Citation2012) with A. acutiloba as the reference (Accession no.: NC_029391), and the annotation was conducted through DOGMA (Wyman et al. Citation2004). Three cp genome varied from 142,484 to 142,529 bp in length, and the overall GC content of each of them was about 37.5%. We predicted 124–126 genes including 81–83 protein-coding genes, 35 tRNA, and 8 rRNA. Annotated chloroplast genomes were deposited in the Genbank (A. sinensis_WMX: MK688991, A. sinensis_WEM: MK688990, A. sinensis_CHQ: MK688989).
To investigate the relationship between the cultivated and wild A. sinensis, we selected 10 complete chloroplast genome sequences for phylogenetic analysis, including A. gigas, A. decursiva, A. nitida, A. dahurica, A. acutiloba, A. tsinlingensis and four individuals from A. sinensis. All sequences were aligned using MAFFT (Katoh and Standley Citation2013). The phylogenetic relationship was reconstructed based on the neighbour-joining (NJ) analysis (Saitou and Nei Citation1987) using MEGA7 (Kumar et al. Citation2016) with 1000 bootstrap replicates. The resulted phylogenetic tree shows that relationships among species of Angelica is consistent with other studies (Zhang et al. Citation2018; Tian et al. Citation2019) (). Four individuals of A. sinensis clustered in one clade, but two wild samples were not separated from the cultivated.
Figure 1. Phylogenetic relationships inferred from 10 chloroplast genomes in Angelica. Bootstrap support is indicated for each node.
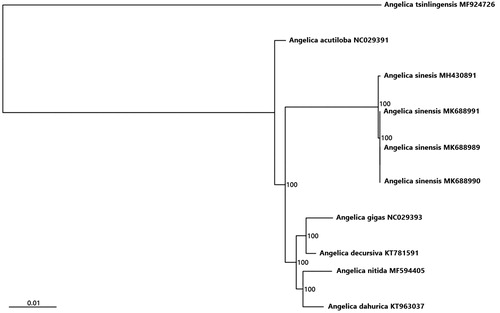
Our studies show that the wild and cultivated samples of A. sinensis share the similar structure on the cp genome and have no significant differentiation on phylogeny, and only 10 variable sites were detected in aligned dataset of four cp genome sequences. Our results seem to indicate that sampled wild A. sinensis should be the escape of the cultivated, not the wild ancestor.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Sun H, Zhang B, Qi Y, Zhang Z, Guo Z. 2009. Survey and analysis on the resource of Angelica sinensis. Chin Agr Sci Bull. 25:437–441.
- Tian E, Liu Q, Chen W, Li F, Chen A, Li C, Chao Z. 2019. Characterization of complete chloroplast genome of Angelica sinensis (Apiaceae), an endemic medical plant to China. Mitochondrial DNA Part B. 4:158–159.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang J, Li D, Li H. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.
- Zhang H, Luo L, Yu Y, Qiu J, Hu Z, Liao W. 2009. The study of the germplasm survey of Angelica sinensis. J Chin Med Mat. 32:335–337.
- Zhang H, Mittal N, Leamy L, Barazani O, Song B. 2017. Back into the wild – apply untapped genetic diversity of wild relatives for crop improvement. Evol Appl. 10:5–24.
- Zhang H, Wang X, Cao D, Niu J, Wang Z. 2018. The complete chloroplast genome sequence of Angelica Tsinlingensis (Apioideae). Mitochondrial DNA Part B. 3:480–481.
