Abstract
In this article, we describe the complete mitochondrial genome of Neotermes koshunensis from the Pingtung County, Taiwan. This mitogenome is 15,589 bp long, containing 13 protein-coding genes, 22 tRNA genes, and 2 rDNA genes. Nucleotide composition of the whole mitogenome is 42.86% for A, 25.42% for T, 19.65% for C, and 12.07% for G. The AT and GC skewness of mitogenomic sequences are 0.255 and −0.239, showing the A-skew and C-skew. Neotermes koshunensis grouped within the clade including the other nine Kalotermitidae species was well supported. The phylogenetic position of Kalotermitidae is sister to Neoisoptera (including Termitidae, Rhinotermitidae, Serritermitidae, and Stylotermitidae) in the current phylogenetic analysis. Mitogenomic data from this study will provide useful information for further studies on the phylogeny of Kalotermitidae.
Termites have been proven to be social cockroaches which are sister to the woodroach Cryptocercus by many comprehensive molecular phylogenetic analyses (Inward et al. Citation2007; Cameron et al. Citation2012; Bourguignon et al. Citation2015). There are 458 species assigning to 23 living genera in termite family Kalotermitidae in the world (Krishna Citation1961; Krishna et al. Citation2013; Ghesini et al. Citation2014; Scheffrahn et al. Citation2018). The habitat of Kalotermitidae is in wood in small colonies which are with irregular galleries filled with faecal pellets. The nests are without connection to soil, except the genus Paraneotermes which forages in soil (Krishna et al. Citation2013). Neotermes koshunensis (Shiraki, 1909) is distributed in China, Japan, Hong Kong, Taiwan, and Vietnam (Krishna et al. Citation2013). This is the first report of complete mitochondrial sequences for the species N. koshunensis.
The single specimen of N. koshunensis in this study was collected in Hengchun, Pingtung County, Taiwan in March 2014. Total genomic DNA was extracted from the soldier’s thorax using the QuickExtract™ DNA Extraction Solution kit (Epicentre, Madison, WI) following the supplier’s instructions. The voucher specimen’s genomic DNA and the partial specimen were deposited in the Taiwan Forestry Research Institute, Taipei, Taiwan. The complete mitogenome of N. koshunensis was sequenced using the next-generation sequencing method (Illumina MiSeq, San Diego, CA) (Hahn et al. Citation2013). A total of 3.6 GB next-generation sequencing paired-end reads were used to assemble the complete mitogenome sequence. The CLC Genomics Workbench (QIAGEN) was used for sequence quality analysis, data trimming, and de novo assembling. The locations of the protein-coding genes, ribosomal RNAs (rRNAs), and transfer RNAs (tRNAs) were predicted by using MITOS Web Server (Bernt et al. Citation2013) and identified by alignment with other mitogenomes of termites. The AT and GC skew was calculated according to the following formulas: AT skew=(A − T)/(A + T) and GC skew=(G – C)/(G + C) (Perna and Kocher Citation1995). The complete mitogenome of N. koshunensis is 15,589 bp in length (GenBank Accession No. MK140662), including 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and one control region. The total nucleotide composition of the N. koshunensis mitogenome was 42.86% for A, 25.42% for T, 19.65% for C, and 12.07% for G. The AT and GC skewness of mitogenome sequence were 0.255 and −0.239, showing the A-skew and C-skew. The skew statistics (AT and GC) of protein-coding gene were 0.277 and −0.221, showing the same A-skew and C-skew. The gene rearrangement of the mitogenome in N. koshunensis is identical to the ancestral insect type (Cameron Citation2014).
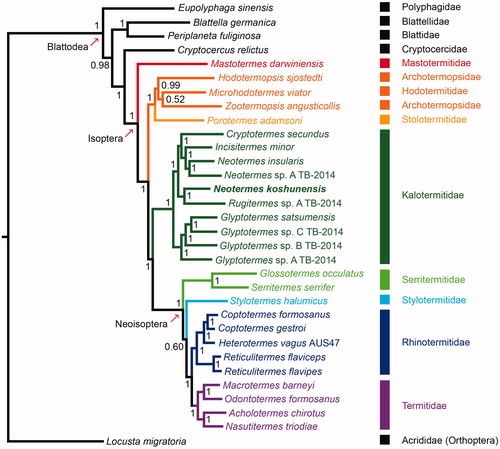
We reconstructed the phylogenetic relationships of 31 Blattodea species and one outgroup (Locusta migratoria) based on 13 mitochondrial protein-coding genes by using Mrbayes version 3.2.4 (Qiagen, Shanghai, China) (Huelsenbeck and Ronquist Citation2001) under model GTR + I+G (). Values on nodes indicated posterior probabilities. N. koshunensis grouped within the clade including the other nine Kalotermitidae species was solid supported (100%). The phylogenetic position of Kalotermitidae is sister to Neoisoptera (including Termitidae, Rhinotermitidae, Serritermitidae, and Stylotermitidae) in the current phylogenetic analysis, consistent with the results of previous studies (Kambhampati et al. Citation1996; Cameron et al. Citation2012; Bourguignon et al. Citation2015). It is obvious that the genus Neotermes is not a monophyletic group based on the present phylogenetic analysis. More complete mitogenomic data of other congeneric species is in need for further studies to clarify the taxonomic status of Neotermes. Mitogenomic data from this study will provide useful information for further studies on the phylogeny of Kalotermitidae.
Figure 1. Phylogenetic tree of the 31 Blattodea species and one Orthoptera species was reconstructed based on the sequences of 13 mitochondrial protein-coding genes with Mrbayes v. 3.2.4 (Huelsenbeck and Ronquist, Citation2001) under model GTR + I+G. Value on nodes indicated posterior probabilities. Acholotermes chirotus (KY224688), Blattella germanica (EU854321), Coptotermes formosanus (AB626145), Coptotermes gestroi (NC_030014), Cryptocercus relictus (JX144941), Cryptotermes secundus (KP026283), Eupolyphaga sinensis (FJ830540), Glossotermes occulatus (KP026291), Glyptotermes satsumensis (KP026257), Glyptotermes sp. A TB-2014 (KP026263), Glyptotermes sp. B TB-2014 (KP026301), Glyptotermes sp. C TB-2014 (KP026300), Heterotermes vagus isolate AUS47 (KU925234)), Hodotermopsis sjostedti (KP026259), Incisitermes minor (MG557850), Locusta migratoria (X80245), Macrotermes barneyi (JX050221), Mastotermes darwiniensis (JX144929), Microhodotermes viator (JX144931), Nasutitermes triodiae (JX144940), Neotermes insularis (JX144933), Neotermes koshunensis(in this study), Neotermes sp. A TB-2014 (KP026299), Odontotermes formosanus (KP026254), Periplaneta fuliginosa (AB126004), Porotermes adamsoni (JX144930), Reticulitermes flaviceps (KX712090), Reticulitermes flavipes (NC_009498) , Rugitermes sp. A TB-2014 (KP026284), Serritermes serrifer (KP026264), Stylotermes halumicus (KY449046), Zootermopsis angusticollis (JX144932).
Acknowledgements
We are grateful to Hsieh Jui-Fan and Jer-Wei Chang for their assistance in DNA data downloading.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bourguignon T, Lo N, Cameron SL, Sobotnik J, Hayashi Y, Shigenobu S, Watanabe D, Roisin Y, Miura T, Evans TA. 2015. The evolutionary history of termites as inferred from 66 mitochondrial genomes. Mol Biol Evol. 32:406–421.
- Cameron SL. 2014. Insect mitochondrial genomics:implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Cameron SL, Lo N, Bourguignon T, Svenson GJ, Evans TA. 2012. A mitochondrial genome phylogeny of termites (Blattodea: Termitoidae): robust support for interfamilial relationships and molecular synapomorphies define major clades. Mol Phylogenet Evol. 65:163–173.
- Ghesini S, Simon D, Marini M. 2014. Kalotermes sinaicus Kemner (Isoptera, Kalotermitidae): new morphological and genetic evidence, and assignment to the new genus Longicaputermes gen. nov. Insectes Soc. 61:123–131.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Inward D, Beccaloni G, Eggleton P. 2007. Death of an order: a comprehensive molecular phylogeneitc study confirms that termites are eusocial cockroaches. Biol Lett. 3:331–335.
- Kambhampati S, Kjer KM, Thorne BL. 1996. Phylogenetic relationship among termite families based on DNA sequence of mitochondrial 16S ribosomal RNA gene. Insect Mol Biol. 5:229–238.
- Krishna K. 1961. A generic revision and phylogenetic study of the family Kalotermitidae (Isoptera). Bull Am Mus Nat His. 122:303–408.
- Krishna K, Grimaldi DA, Krishna V, Engel MS. 2013. Treatise on the Isoptera of the world. Bull Am Mus Nat His. 377:303–408.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41:353–358.
- Scheffrahn RH, Bourguignon T, Akama PD, Sillam-Dussès D, Šobotník J. 2018. Roisinitermes ebogoensis gen. & sp. n., an outstanding drywood termite with snapping soldiers from Cameroon (Isoptera, Kalotermitidae). ZooKeys. 787:91–105.
