Abstract
The complete mitochondrial genome of the box-like thornback cowfish (Lactoria fornasini) was first determined in this study. It was a circular molecule of ∼1.6 kb, including 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and an AT-rich control region. Of the 37 genes, 28 were encoded by the heavy strand, while the rest were encoded by the light strand. The genome composition with A + T bias (56.22%) and gene arrangement were almost identical with those observed in most vertebrates. The 22 tRNA genes ranged from 66 bp (tRNA-Cys) to 75 bp (tRNA-Lys). The phylogenetic result showed that L. fornasini was clustered with L. diaphana.
The thornback cowfish (Lactoria fornasini) (Ostraciidae, Tetraodontiformes) naturally inhabits clear outer lagoon and seaward reefs and appears to be common in Indo-West Pacific and Southeast Atlantic (Smith Citation1986; Myers Citation1999). This hexagonal, box-like fish, can fuse its eyes, mouth, fins, and tail protrude together, to form a solid carapace and in some regions, this fish is poisonous (Svenska Citation1896). In this study, we firstly report the complete mitochondrial DNA genome for L. fornasini and analyzed the phylogenetic relationship of the species in Tetraodontiformes fishes.
The tissue of L. fornasini (voucher No. OKA2016061113) was collected from Okinawa Island, Japan. The experimental protocol and data analysis methods followed Li et al. (Citation2015). Excluding L. fornasini, 20 species of Tetraodontiformes were selected to conduct the phylogenetic tree. The maximum likelihood (ML) method was fulfilled with the AIC model in PhyML 3.0 by two partitions of the mitogenomic data: the first and second codons of the 12 protein-coding genes (except the light-strand encoded ND6 gene) (Guindon et al. Citation2010).
The complete mitogenome of Lactoria fornasini was a classical double-stranded circular molecule which was 16,488 bp in length (GenBank accession no. MK391749). It has a typical mitogenomic organization and gene order as most vertebrates, containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a non-coding control region (D-loop). The overall nucleotide composition of the heavy strand of the L. fornasini mitogenome was as follows: 28.7% A, 27.4% C, 16.4% G, and 27.5% T, with a total A + T content of 56.2%. Similar to the vertebrate mitochondrial genomes, the heavy strand encoded 12 PCGs and 14 tRNA genes, and 2 rRNA genes, whereas the light strand encoded only one PCG and the rest tRNA genes. The 22 tRNA genes ranged from 66 bp (tRNA-Cys) to 75 bp (tRNA-Lys). All tRNAs folded into a conventional cloverleaf shaped secondary structure as identified by tRNAscan-SE (Lowe and Eddy Citation1997). The sizes of the small ribosomal RNA (12S) and large ribosomal RNA (16S) genes were 946 and 1684 bp with an A + T content of 53.6 and 57.2%, respectively. For the 13 PCGs, the COI gene owned a nonstandard initial codon GTG which is common in vertebrates (Slack et al. Citation2003), while the remaining were started by ATG. Two types of termination codon (TAA and TAG) were identified in 10 protein-coding genes, and three incomplete termination codons (T–) were found in the other three genes (COIII, ND4, and Cytb).
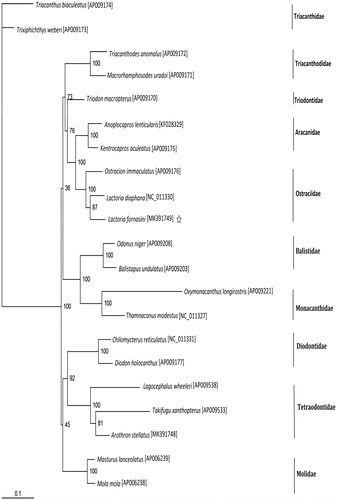
In the Maximum likelihood tree, the result suggested that L. diaphana was most closely related to L. fornasini among all the Tetraodontiformes species included in the analysis. This result is consistent with conventional morphological taxonomy ().
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. System Biol. 59:307–321.
- Li J, Chen X, Kang B, Liu M. 2015. Mitochondrial DNA genomes organization and phylogenetic relationships analysis of eight Anemone fishes (Pomacentridae: Amphiprioninae). PLoS ONE. 10:e0123894.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 25:955–964.
- Myers RF. 1999. Micronesian reef fishes. Barrigada, USA: Coral Graphics.
- Slack KE, Janke A, Penny D, Arnason U. 2003. Two new avian mitochondrial genomes (penguin and goose) and a summary of bird and reptile mitogenomic features. Gene. 302:43–52.
- Smith MM. 1986. Smiths’ sea fishes. Berlin, Germany: Springer-Verlag.
- Svenska A. 1896. The fishes of The Indo-Australian Archipelago, Vol XI. Leiden, the Nethelands: Brill Publish.