Abstract
Paulownia elongata (Paulowniaceae) is an ornamental tree native to Asia. In this study, the chloroplast (cp) genome of P. elongata was assembled using genome skimming sequencing, and the phylogeny of Lamiales was reconstructed based on whole cp genome data. The cp genome of P. elongata is 154,670 bp in length, comprising two copies of inverted region (IR, 25,783 bp), regions separated by the large single copy (LSC, 85,370 bp) and small single copy (SSC, 17,734 bp) regions. The cp genome encodes 114 unique genes, including 80 different protein-coding genes, 30 tRNA genes, and four rRNA genes, with 19 duplicated genes in the IR regions. Phylogenetic analysis indicates that P. elongata together with two other Paulownia species form a clade, which is sister to Triaenophora shennongjiaensis.
Paulownia Sieb. et Zucc. is known as an economically important multipurpose tree genus due to its fast growth and short-rotation harvesting for timber (Stewart et al. Citation2018). There are seven species native to China in this genus (Wu and Raven Citation2000). Among these species, Paulownia elongata is a widely distributed tree native to Asia and known for its high adaptability and rapid growth rate (Chaires et al. Citation2017). In addition, P. elongata is also highly tolerant to a wide variety of biotic and abiotic stresses (Chaires et al. Citation2017). However, information on the genetic background of P. elongata remains scarce. In this study, the complete chloroplast (cp) genome of P. elongata was determined using genome skimming data. This genome sequence was deposited into GenBank (accession number MK805127).
Total genomic DNA was isolated from fresh leaf tissue of one P. elongata plant collected in Lankao (China; 114°50′40.33″E, 34°50′39.84″N) using Plant DNAzol Reagent (LifeFeng, Shanghai) according to the manufacturer’s protocol. A voucher specimen (Haodi Zhao ZHD181105) was deposited at the Herbarium of Zhejiang University (HZU). The high quality DNA was sheared and the paired-end libraries preparation and sequencing were performed on Illumina HiSeq 2500 by Beijing Genomics Institute (Wuhan, China). The raw data were filtered by quality with Phred score <30. All remaining sequences were assembled into contigs using CLC de novo assembler beta 4.06 (CLC Inc., Rarhus, Denmark) to reconstruct the cp genome, with P. coreana (GenBank accession number: KP718622) as a reference. The annotation of the cp genome was performed through the online program Dual Organellar Genome Annotator (Wyman et al. Citation2004), and the circular gene maps were drawn using the OrganellarGenomeDRAW tool (OGDRAW) as described in Liu et al. (Citation2017). Phylogenetic tree for 22 whole cp genome sequences of Lamiales was constructed using maximum likelihood (ML) and Bayesian inference (BI) methods, with Tiquilia plicata as outgroup. ML analysis was carried out using RAxML-HPC v8.1.11 on the CIPRES cluster (Miller et al. Citation2010) and BI analysis was performed using MrBayes v3.2.3 (Ronquist and Huelsenbeck Citation2003).
The complete cp genome of P. elongata is 154,670 in length and, similar to those of other flowring plants, comprises two copies of IR (25,783 bp) regions separated by the LSC (85,370 bp) and SSC (17,734 bp) regions. Within the cp genome of P. elongata, there are 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes, with additional 19 duplicated genes in the IR regions. Six tRNA genes and nine protein-coding genes contain a single intron, and three genes (i.e., rps12, clpP, and ycf3) contain two introns. The constructed phylogeny showed that three Paulownia species, including P. tomentosa, P. coreana, and P. elongata, formed a highly supported clade, which in turn was sister to Triaenophora shennongjiaensis (Orobanchaceae) ().
Figure 1. Phylogenetic relationships of Lamiales inferred based on whole cp genome sequences. Relative branch lengths are indicated at the top-left corner. Numbers above the branches represent bootstrap values from maximum-likelihood analyses and posterior probabilities from Bayesian inference, respectively. GenBank accession numbers of taxa are shown below, Paulownia tomeniosa (KP718624), Paulownia coreana (KP718622), Triaenophora shennongjiaensis (MH071405), Erythranthe lutea (KU705476), Scutellaria baicalensis (MF521633), Premna microphylla (KM981744), Adenocalymma nodosum (MG831862), Tanaecium tetragonolobum (KR534325), Aloysia citrodora (KY085903), Sesamum indicum (JN637766). Utricularia foliosa (KY025562), Genlisea violacea (MF593126), Andrographis paniculata (KF150644), Echinacanthus lofouensis (MF490441), Scrophularia henryi (MF861203), Veronica nakaiana (KT633216), Veronicastrum sibiricum (KT724053), Primulina linearifolia (MF472013), Boea hygrometrica (JN107811), Chionanthus retusus (KY582962), Forsythia suspensa (MF579702), and Tiquilia plicata (MG573056).
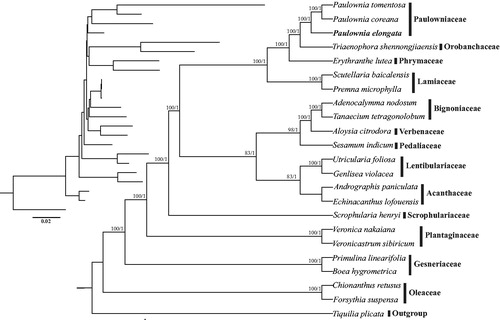
In conclusion, the complete cp genome of P. elongata is reported for the first time in this study. It will provide not only essential and important genetic resources for future better development and utilization of this species but also insights into evolutionary relationships within the Order Lamiales.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chaires M, Gupta D, Joshee N, Cooper KK, Basu C. 2017. RNA-seq analysis of the salt stress-induced transcripts in fast-growing bioenergy tree, Paulownia elongata. J Plant Interact. 12:128–136.
- Liu LX, Li R, James RP, Worth Li X, Li P, KM, Cameron, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968
- Miller M-A, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010. IEEE. p 1–8.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stewart WM, Vaidya BN, Mahapatra AK, Terrill TH, Joshee N. 2018. Potential use of multipurpose Paulownia elongata tree as an animal feed resource. Am J Plant Sci. 09:1212.
- Wyman S-K, Jansen R-K, Boore J-L. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Wu ZY, Raven PH. 2000. Flora of China, Vol. 18: Scrophulariaceae through Gesneriaceae. Beijing: Science Press and St Louis: Missouri Botanical Garden Press.
