Abstract
In this paper, we sequenced the complete mitochondrial genome of Menida violacea. It was 15,787 bp in length, with a base composition of 41.83% A, 33.43% T, 13.82% C, and 10.93% G. It contained 37 typical mitochondrial genes (13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region). Two kinds of the initiation codon and three kinds of termination codon were employed in the 13 PCGs. Phylogenetic analysis implied that M. violacea belongs to the family Pentatomidae, and each family in Pentatomoidea formed a monophyletic cluster with a high degree of bootstrap support.
Menida violacea Motschulsky, 1861(Hemiptera: Pentatomidae) is an agricultural pest widely distributed in China, the eastern Siberia of the former Soviet Union, Japan and other places (Li et al. Citation2015). It is harmful to crops such as rice, corn, wheat and trees such as apple, pear, and elm (Yang Citation1962). In view of the importance of M. violacea, we sequenced its complete mitochondrial genome and used this to clarify its phylogenetic position.
In this study, the adult samples were collected from Huanghualing in Niubeiliang Nature Reserve (33.7°N, 108.8°E, alt. 1299 m), Shaanxi Province, China. The voucher specimens and the remaining genomic DNA were preserved in the Institute of Entomology, Shanxi Agricultural University. The complete mitochondrial genome of M. violacea was a double-stranded covalently closed circular DNA molecule, with a total length of 15,787 bp (GenBank accession number: MK617948) and containing 13 protein-coding genes (PCGs), 22 tRNA genes (tRNAs), 2 rRNA genes (rrnL and rrnS), and a control region. Among them, 23 genes were located on J-strand, and the other genes (ND5, ND4, ND4L, ND1, trnQ, trnC, trnY, trnF, trnH, trnP, trnL(CUN), trnV, 12S rRNA, and 16S rRNA) were located on the N-strand. The gene order and orientation of the mitochondrial genes were identical to those of most Pentatomidae species (Yuan et al. Citation2015, Zhao et al. Citation2018).
The nucleotide composition of the M. violacea mitogenome was significantly biased towards A and T, with a 75.25% A + T content on average. Most PCGs started with ATN codons, except COI, ATP8, and ND6, which started with TTG. 10 PCGs terminated with TAA, except ND4 ended with TAG, two PCGs (COII and ND3) terminated with a single T. The length of 22 tRNAs range from 64 to 71 bp, and had a typical cloverleaf secondary structure except trnS-Ser(GCT) and trnV-Val (TAC), which lack a dihydrouridine arm. The lengths of two rRNAs were 803 bp (12S rRNA) and 1284 bp (16S rRNA).
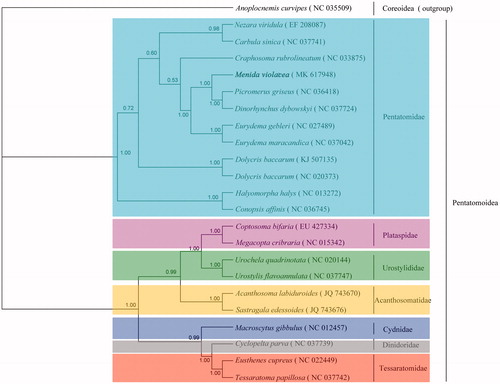
Phylogenetic analyses of 23 Pentatomoidea species were based on Bayesian inference (Zhou et al. Citation2011) using the concatenated nucleotide sequences of 13 mitochondrial PCGs. In the result, each family formed a monophyletic cluster with a high degree of bootstrap support (), and it was identical to the previous study (Zhao et al. Citation2017). The species M. violacea was situated in the clade of Pentatomidae. While in the Pentatomidae, the subfamilies were mixed with each other and did not have a clear phylogenetic relationship. This implied that further studies are needed and more species needed to be sequenced in Pentatomidae, which will enhance our understanding of molecular phylogeny in Pentatomidae. The complete mitogenome of M. violacea provides important molecular and evolutionary evidence for Pentatomidae and would play an important role in further phylogeny systematic studies.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Li XR, Fan ZH, Liu GQ. 2015. Note on genus Menida motschulsky from China (Hemiptera: Pentatomidae). J Tianjin Normal Univ (Natural Science Education). 35:12–22.
- Yang WI. 1962. Hemiptera, Pentatomidae. In: Economic insect fauna of China. vol. 2. Beijing, China: Academia Sinica, Science Press; p. 138.
- Yuan ML, Zhang QL, Guo ZL, Wang J, Shen YY. 2015. Comparative mitogenomic analysis of the superfamily Pentatomoidea (Insecta: Hemiptera: Heteroptera) and phylogenetic implications. BMC Genomics. 16:460.
- Zhao Q, Wang J, Wang MQ, Cai B, Zhang HF, Wei JF. 2018. Complete mitochondrial genome of Dinorhynchus dybowskyi (Hemiptera: Pentatomidae: Asopinae) and phylogenetic analysis of pentatomomorpha species. J Insect Sci. 18:44.
- Zhao WQ, Zhao Q, Li M, Wei JF, Zhang XH, Zhang HF. 2017. Characterization of the complete mitochondrial genome and phylogenetic implications for Eurydema maracandica (Hemiptera: Pentatomidae). Mitochondrial DNA B Resour. 2:550–551.
- Zhou JF, Liu XG, Stones DS, Xie Q, Wang G. 2011. MrBayes on a graphics processing unit. Bioinformatics. 27:1255–1261.