Abstract
The complete chloroplast genome of Aesculus chinensis was obtained with Illumina HiSeq X Ten. The chloroplast genome is 155,528 bp in length, including a pair of inverted repeat (IR) regions of 25,656 bp, a large single-copy (LSC) region of 85,489 bp, and a small single-copy (SSC) region of 18,727 bp. It contains 115 genes, including 80 protein-coding genes, 31 transfer RNAs, and four ribosomal RNAs. The total GC content is 37.9%, whereas the corresponding values of the LSC, SSC, and IR regions are 36.1%, 31.8%, and 43.2%, separately. Phylogenetic analysis of the protein-coding genes showed a close relationship with Aesculus wangii in Hippocastanaceae.
Aesculus chinensis (Hippocastanaceae), abundant in the northwestern China, is a widely used medicinal plant rich of escins (Cheng et al. Citation2018). Modern pharmacological studies show that A. chinensis possess diverse biologic activities such as antiviral (Yang et al. Citation1999), anti-inflammatory (Zhao et al. Citation2001; Wei et al. Citation2004), antitumor (Wang et al. Citation2017), antioxidative (Sato et al. Citation2005; Cheng et al. Citation2018), and antigenotoxic properties (Sato et al. Citation2005; Cheng et al. Citation2018). Its dried ripe seeds have been used as a stomachic and analgesic in the treatment of ditension and pain in the chest and abdomen (Wei et al. Citation2004). Aesculus chinensis has high prospects to be further developed for tree cultivation.
The chloroplast genome sequences have been utilized as reliable tools for phylogeneitc and evolutionary research (Wang et al. Citation2018). In recently, many chloroplast genomes of valuable plants have been reported (Chen et al. Citation2018; Liu and Han, Citation2018; Zheng et al. Citation2018; Wang et al. Citation2018; Kwon et al. Citation2019; Yang et al. Citation2019; Zhu and Sun Citation2019). To facilitate its genetic research and contribute to its utilization, we reported the complete chloroplast genome sequence of A. chinensis based on Illumina HiSeq X Ten. The annotated genomic sequence has been submitted to GenBank with the accession number MK737939. Phylogenic evolution analysis revealed that its phylogenetic placement within the family Hippocastanaceae.
The pattern specimens were taken from Shaanxi Province in Southwest of China, located at 105°59′40″E, 33°11′57″N and were stored in the Herbarium of Neijiang Normal University (accession number: 20190122AC02). Chloroplast DNA (cpDNA) extracted following the directions of DNeasy Plant Mini Kit (Qiagen, CA, USA). The whole genome sequencing was conducted on the Illumina HiSeq X Ten Sequencing System in Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The complete chloroplast genome was assembled using the baiting and iterative mapping approach (Hahn et al. Citation2013), with that of its congener Aesculus wangii (GenBank: MF583747) as the initial reference genome. A total of 4,323,520 individual chloroplast reads yielded an average coverage of 141.3-fold. The chloroplast genome was annotated in GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand) and was drawn to the circular chloroplast genome sequence map of OGDRAW.
The chloroplast genome of A. chinensis is 155,528 bp in length (GenBank accession number: MK737939) and exhibits a typical quadripartite structure of the large (LSC, 85,489 bp) and small (SSC, 18,727 bp) single-copy regions, separated by a pair of inverted repeat regions (IRs, 25,656 bp each). The total GC content is 37.9%, whereas the corresponding values of the LSC, SSC, and IR regions are 36.1%, 31.8%, and 43.2%, respectively. The complete chloroplast genome of A. chinensis contains 115 unique genes, including 80 protein-coding genes, 31 transfer RNA genes, and four ribosomal RNA genes. Intron-exon structure analysis indicated that 97 (84.3%) genes had no introns, whereas 16 (13.9%) genes contained a single intron and two protein-coding genes had two introns.
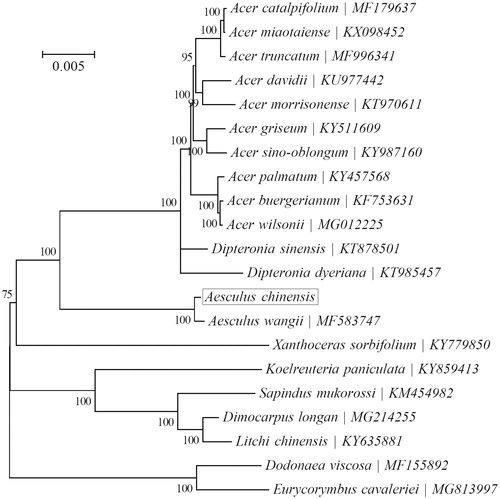
In order to verify the evolutionary relationship, maximum-likelihood phylogenetic tree was generated using the whole chloroplast DNA alignment from 20 published plant species in Sapindaceae using MEGA 7.0 (Kumar et al. Citation2016), which showed that A. chinensis has a close relationship with Aesculus wangii in Hippocastanaceae (). The results will provide a foundation for further investigation of chloroplast genome evolution in Hippocastanaceae.
Figure 1. Phylogeny of 21 species within the family Sapindaceae based on the maximum-likelihood (ML) of the concatenated chloroplast protein-coding sequences. The GTR + G + I model was employed as the best-fit nucleotide substitution model as suggested using MEGA7. The support values are based on 100 bootstrap replicates and are placed next to the branches.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen L, Li L, Yang G, Li M. 2018. Characterization of the complete chloro-plast genome sequence of Tsuga longibracteata WC Cheng (Pinaceae). Conserv Genet Resour. 11:117–120.
- Cheng JT, Chen ST, Guo C, Jiao MJ, Cui WJ, Wang SH, Deng Z, Chen C, Chen S, Zhang J, Liu A. 2018. Triterpenoid saponins from the seeds of Aesculus chinensis and their cytotoxicities. Nat Prod Bioprospect. 8:47–56.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucl Acids Res. 41:e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Kwon W, Kim Y, Park CH, Park J. 2019. The complete chloroplast genome sequence of traditional medical herb, Plantagodepressa Willd. (Plantaginaceae). Mitochondrial DNA B. 4:437–438.
- Liu Y, Han Y. 2018. The complete chloroplast genome sequence of Camellias (Camellia fangchengensis). Mitochondrial DNA B. 3:34–35.
- Sato I, Suzuki T, Kobayashi H, Tsuda S. 2005. Antioxidative and antigenotoxic effects of Japanese horse chestnut (Aesculus turbinata) seeds. J Vet Med Sci. 7:731–734.
- Wang M, Wang C, Zhang M, Wang Y, Liu M, Liu Z. 2017. Study on production of escin by embryos of Aesculus chinensis cell. J Chinese Med Mater. 7:1552–1557.
- Wang L, Sui C, Wang Y, Guo J. 2018. The complete chloroplast genome of the endangered plant Paphiopedilum wardii (Orchidaceae). Mitochondrial DNA B. 3:1256–1258.
- Wei F, Ma LY, Jin WT, Ma SC, Han GZ, Khan IA, Lin RC. 2004. Antiinflammatory triterpenoid saponins from the seeds of Aesculus chinensis. Chem Pharm Bull (Tokyo). 10:1246–1248.
- Yang X, Qian X, Wang Z. 2019. The complete chloroplast genome of Casuarina glauca. Mitochondrial DNA B. 4:357–358.
- Yang XW, Zhao J, Cui YX, Liu XH, Ma CM, Hattori M, Zhang LH. 1999. Anti-HIV-1 protease triterpenoid saponins from the seeds of Aesculus chinensis. J Nat Prod. 62:1510–1513.
- Zhao J, Yang XW, Hattori M. 2001. Three new triterpene saponins from the seeds of Aesculus chinensis. Chem Pharm Bull. 5:626–628.
- Zheng W, Wang W, Harris AJ, Xu X. 2018. The complete chloroplast genome of vulnerable Aesculus wangii (Sapindaceae), a narrowly endemic tree in Yunnan, China. Conservation Genet Resour. 10:335–338.
- Zhu X, Sun Y. 2019. The complete chloroplast genome of the endangered tree Parashorea chinensis (Dipterocarpaceae). Mitochondrial DNA B. 4:1163–1164.
