Abstract
The complete plastome sequence of Microlicia cogniauxiana R. Romero (Melastomataceae) (NCBI accession number MK726004), a plant endemic to the campos rupestres formations of the Brazilian savannahs, is presented in this study. A genomic unit is 155,883 bp in length, comprising 130 genes, 85 DNA coding sequences, 37 tRNAs, eight rRNAs, and 23 introns, and a GC content of 37%. We recovered a large single-copy (85,691 bp), a small single-copy (16,859 bp), and two inverted repeats (26,667 bp). The M. cogniauxiana plastome resembles the ancestral organization of plastomes of flowering plants, including those of other genera of Melastomataceae.
Microlicia D. Don is the largest genus in the tribe Microlicieae Naudin with over 150 species (Flora do Brasil 2020 em construção Citation2019). The majority of the species in the genus occur in campos rupestres vegetative formation in the Brazilian savannahs (Romero and Martins Citation2002), a biome considered one of the world’s biodiversity hotspots (Myers et al. Citation2000). Microlicia cogniauxiana R. Romero is endemic to the southern portion of the Espinhaço Range (Minas Gerais, Brazil), occurring in patches of sandy soil among rocks (Romero et al. Citation2015). To date, plastome sequences of 19 other species of Melastomataceae are available on GenBank (Reginato et al. Citation2016; He et al. Citation2017). Here, we present the first complete plastome of the genus Microlicia (NCBI accession number MK726004) as a resource for future investigations.
A voucher specimen (Romero et al. 8650, HUFU70585) was deposited in the Herbarium Uberlandense from the Federal University of Uberlândia (HUFU). Genomic DNA (gDNA) was isolated from dried leaf tissue using the Qiagen DNeasy Plant Mini Kit. Total gDNA was sequenced using libraries with insert sizes of ca. 300 bp and Illumina HiSeq 2500 (Illumina Inc., San Diego, CA). A total of 2,424,834 paired-end reads were quality trimmed and assembled de novo using BBTools (available at https://jgi.doe.gov/data-and-tools/bbtools/) and Ray v. 2.3.4 (Boisvert et al. Citation2012). Annotation was done in Chlorobox GeSeq (Tillich et al. Citation2017) and confirmed in Geneious v. 7.1.9 (Kearse et al. Citation2012). Alignment of 78 plastid protein-coding genes was done in MAFFT v. 7.309 (Katoh et al. Citation2017). A phylogenetic reconstruction of 20 species of Melastomataceae for which plastome sequences were available was performed using a maximum-likelihood framework in IQ-TREE v. 1.5.5 (Nguyen et al. Citation2015).
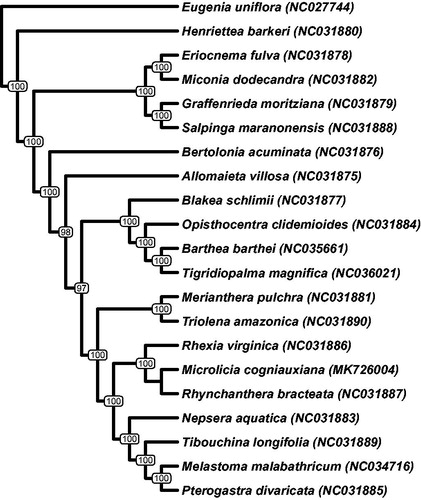
The plastome monomer has a typical four-partite structure in which a single monomer is 155,883 bp in length with a large single copy of 85,691 bp, a small single-copy of 16,859 bp, and two inverted repeats of 26,667 bp. Microlicia cogniauxiana’s plastome structure, gene order, and gene content resembles the plastome of other species of Melastomataceae. The phylogeny inferred with 78 plastid protein-coding genes is concordant with the current understanding of the Melastomataceae relationships (Reginato et al. Citation2016; He et al. Citation2017). Microlicieae encompasses Microlicia and six other genera; Chaetostoma DC., Lavoiseira DC., Trembleya DC., Stenodon Naudin, Poteranthera Bong., and Rhynchanthera DC. (Almeda and Martins Citation2001; Fritsch et al. Citation2004). The monophyly of this tribe is supported by previous molecular and morphological data (Fritsch et al. Citation2004; Michelangeli et al. Citation2013; Rocha et al. Citation2016) and was also supported in this study by the sister relationship between M. cogniauxiana and Rhynchanthera bracteata ().
Figure 1. Maximum likelihood phylogeny representing the relationship of Microlicia cogniauxiana and the 19 other species of Melastomataceae for which plastomes are available on GenBank. Phylogeny was inferred with 78 plastid protein-coding genes using GTR + I + G and 1000 bootstrap replicates with best tree replicate log-likelihood of -246361.8409. Eugenia uniflora (Myrtaceae) was used as outgroup.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for the content and writing of the paper.
Additional information
Funding
References
- Almeda F, Martins AB. 2001. New combinations and new names in some Brazilian Microlicieae (Melastomataceae), with notes on the delimitation of Lavoisiera, Microlicia and Trembleya. Novon. 11:1–7.
- Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. 2012. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol. 13:1–13.
- Flora do Brasil 2020 em construção. 2019. Jardim Botânico do Rio de Janeiro. Available at <http://floradobrasil.jbrj.gov.br/> [accessed 2019 February 21].
- Fritsch PW, Almeda F, Renner SS, Martins AB, Cruz BC. 2004. Phylogeny and circumscription of the near-endemic Brazilian tribe Microlicieae (Melastomataceae). Am J Bot. 91:1105–1114.
- He X, Zhou Y, Cai Y, Chen Z, Wu W, Zhou R, Ng WL. 2017. The complete chloroplast genome sequence of Barthea barthei (Melastomataceae), a shrub endemic to southern China. Mitochondrial DNA B. 2:810–811.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 1–7.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Michelangeli FA, Guimarães PJF, Penneys DS, Almeda F, Kriebel R. 2013. Phylogenetic relationships and distribution of New World Melastomeae (Melastomataceae). Bot J Linn Soc. 171:38–60.
- Myers N, Fonseca GAB, Mittermeier RA, Fonseca GAB, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature. 403:853–858.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Reginato M, Neubig KM, Majure LC, Michelangeli FA. 2016. The first complete plastid genomes of Melastomataceae are highly structurally conserved. PeerJ. 4:e2715.
- Rocha MJRD, Guimarães PJF, Michelangeli FA, Romero R. 2016. Phylogenetic placement and a new circumscription of Poteranthera (Microlicieae; Melastomataceae). Phytotaxa. 263:219–232.
- Romero R, Martins AB. 2002. Melastomataceae do Parque Nacional da Serra da Canastra, Minas Gerais, Brasil. Rev Brasil Bot. 25:19–24.
- Romero R, Silva KR, Simão DG. 2015. Microlicia cogniauxiana and Microlicia naudiniana (Melastomataceae), two new species from the Espinhaço Range, Brazil. Syst Bot. 40:1012–−1021.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
