Abstract
The complete chloroplast genome of Conyza canadensis (GenBank accession number: MK737940) was obtained by using Illumina HiSeq X Ten in this study. The circular Conyza canadensis chloroplast genome is 152,721 bp long and comprises of a pair of inverted repeat (IR) regions of 25,004 bp each, a large single-copy (LSC) region of 84,280 bp, and a small single-copy (SSC) region of 18,433 bp. The complete chloroplast genome contains 114 unique genes, including 80 protein-coding genes, 30 tRNAs, and 4rRNAs. Phylogenetic analysis of the protein-coding genes revealed that Conyza canadensis was closely related to Conyza bonariensis.
Conyza canadensis, belonging to the family Asteraceae, is commonly known as Canada fleabane, bitterweed, and horseweed, etc. It is an annual medicinal herb that is distributed throughout the world (Shakirullah et al. Citation2011; Phuong et al. Citation2015). It also can be used as a traditional vegetable and a sweetening agent (Shakirullah et al. Citation2011). The plant is full of essential oil containing bioactive compounds. These compounds were identified with multiple biological functions such as antifungal (Veres et al. Citation2012; Phuong et al. Citation2015), anti-inflammatory (Lenfeld et al. Citation1986; Meric Citation2009), antimicrobial and insecticidal (Shakirullah et al. Citation2011; Zeng et al. Citation2014), antiproliferative (Csupor-Löffler et al. Citation2011), anti-melanoma (Yan et al. Citation2010) and antioxidation activity (Hayet et al. Citation2009). The publication of the nuclear genome for C. Canadensis has been available online, but no complete chloroplast sequences have been made available for this genus on public databases (Peng et al. Citation2014; Hereward et al. Citation2017).
The chloroplast genome sequences have been utilized as reliable tools for phylogenetic and evolutionary research (Shi et al. Citation2012; Cheng et al. Citation2018; Wang et al. Citation2018). In this study, we conducted the sequencing of C. canadensis chloroplast genome and the phylogenetic analysis with other Asteraceae chloroplast genomes.
The pattern specimens of C. canadensis were taken from Gansu Province in Southwest of China, located at 105°23′3″E, 33°32′2″N, and were stored in the Herbarium of Neijiang Normal University (accession number: 20190120CC01). Chloroplast DNA (cpDNA) extracted from young and fresh leaves following the directions of the DNeasy Plant Mini Kit (Qiagen, CA, USA). Pure cpDNA were sequenced on the Illumina HiSeq X Ten platform at the Novogene Bioinformatics Technology Co., Ltd., Beijing, China. The complete chloroplast genome was assembled using the baiting and iterative mapping approach (Hahn et al. Citation2013), with that of its congener Conyza bonariensis (GenBankaccession number:MF276802) (Hereward et al. Citation2017) as the initial reference genome. The chloroplast genome was annotated using a web-based annotation program GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) and editing by manual and imagining with OGDraw v1.2 (Lohse et al. Citation2013). The deposited GenBank accession number of C. canadensis is MK737940.
The complete chloroplast genome of C. canadensis was 152,721 bp with a typical circular structure which comprises an LSC region of 84,280 bp, an SSC region of 18,433 bp, and a pair of IR regions (IRa and IRb) of 25,004 bp. The chloroplast genome contained a total of 114 genes including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The GC content of the whole genome is 37.2%, while the corresponding values of the LSC, SSC, and IR regions are 35, 30.9, and 43%, respectively. In addition, among the annotated chloroplast genomic sequence, 14 genes possessed only single intron while 2 genes (clpP and ycf3) possessed two introns.
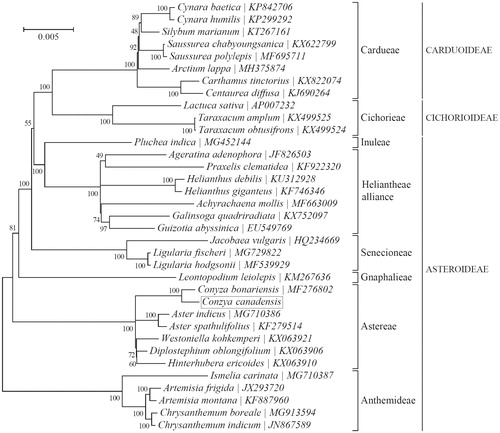
To identify the phylogenetic position of C. canadensis, phylogenetic analysis was conducted with the complete chloroplast genome sequence of C. canadensis and other 34 species from Asteraceae using maximum-likelihood (ML) method in MEGA7 (Kumar et al. Citation2016). The phylogenetic tree () indicated that C. canadensis and the other 6 species were clustered into a clade. In addition, C. canadensis was most closely related to C. bonariensis belonging to the subfamily Astereae.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cheng T, Weng Y, Yang L, Lu L, Hao Z, Shi J, Chen J. 2018. The chloroplast genome of Cerasus campanulata (Maxim.) A.N. Vassiljeva. Mitochondrial DNA Part B. 1:222–224.
- Csupor-Löffler B, Hajdú Z, Zupkó I, Molnár J, Forgo P, Vasas A, Kele Z, Hohmann J. 2011. Antiproliferative constituents of the roots of Conyza canadensis. Planta Medica. 11:1183–1188.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucl Acids Res. 41: e129.
- Hayet E, Maha M, Samia A, Ali MM, Souhir B, Abderaouf K, Mighri Z, Mahjoub A. 2009. Antibacterial, antioxidant and cytotoxic activities of extracts of Conyza canadensis (L.) Cronquist growing in Tunisia. Med Chem Res. 18:447–454.
- Hereward JP, Werth JA, Thornby DF, Keenan M, Chauhan BS, Walter GH. 2017. Complete chloroplast genome of glyphosate resistant Conyzabonariensis (L.) Cronquist from Australia. Mitochondrial DNA Part B. 2:444–445.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lenfeld J, Motl O, Trka A. 1986. Anti-inflammatory activity of extracts from Conyza canadensis. Die Pharmazie. 4:268–269.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and vsualizing expression data sets. Nucl Acids Res. 41:575–581.
- Meric C. 2009. Calcium oxalate crystals in some species of the tribe Inuleae (Asteraceae). Acta Biologica Cracoviensia Series Botanica. 1:105–110.
- Peng Y, Lai Z, Lane T, Nageswara-Rao M, Okada M, Jasieniuk M, O'Geen H, Kim RW, Sammons RD, Rieseberg LH, et al. 2014. De novo genome assembly of the economically important weed horseweed using integrated data from multiple sequencing platforms. Plant Physiol. 166:1241–1254.
- Phuong NB, Lien NTT, Hoai NTT. 2015. Antifungal activity of Conyza canadensis ((L.) Cronquist) collected in northern Viet Nam. In: Proceedings of 5th International Conference on Biomedical Engineering in Vietnam; June 16–18; Vietnam. Cham, Switzerland: Springer. p. 359–361.
- Shakirullah M, Ahmad H, Shah MR, Ahmad I, Ishaq M, Khan N, Badshah A, Khan I. 2011. Antimicrobial activities of Conyzolide and Conyzoflavone from Conyza canadensis. J Enz Inhibition and Med Chem. 4:468–471.
- Shi C, Hu N, Huang H, Gao J, Zhao Y-J, Gao L-Z. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One. 7:e31468.
- Wang L, Sui C, Wang Y, Guo J. 2018. The complete chloroplast genome of the endangered plant Paphiopedilum wardii (Orchidaceae). MitochondrialDNA Part B, 2:1256–1258.
- Veres K, Csupor-Löffler B, Lázár A, Hohmann J. 2012. Antifungal activity and composition of essential oils of Conyzacanadensis herbs and roots[J]. The Scientific World Journal. 2012:1.
- Yan MM, Li TY, Zhao DQ, Shao S, Bi SN. 2010. A new derivative of triterpene with anti-melanoma B16 activity from Conyza canadensis. Chin Chem Lett. 21:834–837.
- Zeng DQ, Peng YH, Chen FF, et al. 2014. Insecticidal activity of essential oil derived from horseweed Conyza canadensis (L.) Cronq. against two mosquitoes and its volatile components. Acta Entomologica Sinica. 2:204–211.