Abstract
Chamaesium paradoxum H. Wolff is an endemic species naturally distributed in China. The complete chloroplast genome sequence of C. paradoxum was generated by de novo assembly using whole genome next generation sequencing data. The complete chloroplast genome of C. paradoxum is 153,512 bp in length, consisting of a pair of inverted repeats (IRs, 25,987 bp) separated by a large single-copy region (LSC, 84,162 bp) and a small single-copy region (SSC, 17,376 bp). There are 129 genes annotated, including 84 coding genes, 37 transfer RNA genes (tRNA), and eight ribosomal RNA genes (rRNA).
Chamaesium paradoxum H. Wolff belongs to the genus Chamaesium in Apiaceae (Apioideae) and is a biennial herb grown in the humid environments at an altitude of 3000–5000 m (Pan and She Citation1995). As a species distributed on the Qinghai-Tibet plateau, C. paradoxum is also used as a medicinal resource by the Tibetan people to treat diseases (Zhong et al. Citation2008). The rhizome of C. paradoxum has been used for the treatment of detumescence and relieving and eliminating mass, and the fruit of it has effect on dispelling coldness (Zhong et al. Citation2016). Furthermore, based on previous phylogenetic analyses of molecular data, Chamaesium occupied an isolated position and all Chinese species represent a monophyly based on ITS and cpDNA evidence involved nine species of Chamaesium (Guo et al. Citation2018). However, so far, no complete chloroplast genome has been sequenced and assembled in genus Chamaesium. In this study, the complete chloroplast genome of C. paradoxum is first reported, which is valuable for designing molecular markers and gaining a better insight into phylogenetic relationships, divergence dating, and population genetics.
Mature leaves of C. paradoxum were collected from Kangding county (32°07′N, 101°80′E), Sichuan Province, China, and the voucher specimen (voucher number: GXL15092501) was deposited in Sichuan University Herbarium (SZ). The complete chloroplast genome sequence was deposited in GenBank under accession No: MK780227. Total DNA was extracted using the Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China). The sample of C. paradoxum was sequenced as the paired-end using the Illumina Novaseq 6000 platform (Illumina, San Diego, CA). FastQC (v0.11.8) was used to conduct quality assessment and all clean data was used for de novo assembly using NOVOplasty2.7.2 (Dierckxsens et al. Citation2017). Annotations of chloroplast genome were conducted by the software Geneious v11.0.4 (Kearse et al.Citation 2012). Start/stop codons and intron/exon borders were edited manually after comparison with reference.
The complete chloroplast genome of C. paradoxum exhibited a single circular molecule with 153,512 bp in length. The cp genome contains a pair of identical IRs (25,987 bp) separated by LSC (84,162 bp) and SSC (17,376 bp) regions. The overall GC content was 38.4%, whereas that of the LSC, SSC, and IR region are 36.7%, 32.6%, and 43.1%, respectively. There is a total of 129 genes in the cp genome, including 84 coding genes, 37 transfer RNA genes (tRNA), and eight ribosomal RNA genes (rRNA).
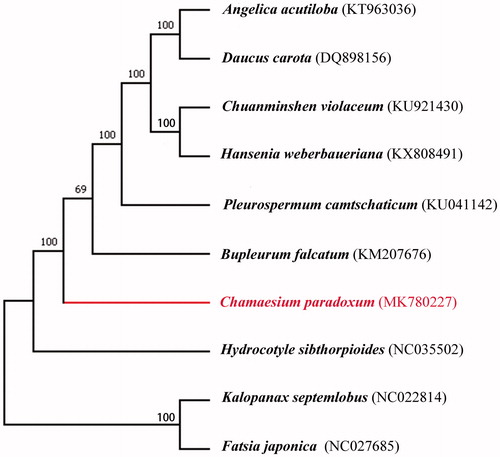
We selected other nine related complete chloroplast genomes from GenBank to assess the phylogenetic relationship with C. paradoxum. The alignment was performed using software MAFFT (Kazutaka and Standley Citation2013). A maximum-likelihood (ML) tree was generated by MEGA7.0 (Kumar et al. Citation2016) using 1000 bootstrap replicates. The phylogenetic tree () indicated that Chamaesium occupied a basal position within the subfamily Apioideae, not the genus Bupleurum, which is inconsistent with the previous studies (Downie et al. Citation2010).
Acknowledgements
The authors are grateful to the opened raw genome data from public database. The authors thank Danmei Su, Xin Yang for the help of sequence analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18
- Downie SR, Spalik K, Katz-Downie DS, Reduron JP. 2010. Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nr DNA ITS sequences. Plant Divers Evol. 128:111–136.
- Guo XL, Wang CB, Wen J, Zhou SD, He XJ. 2018. Phylogeny of Chinese Chamaesium (Apiaceae: Apioideae) inferred from ITS, cpDNA and morphological characters. Phytotaxa. 376:1–016.
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse, M, Moir, R, Wilson, A, Stones-Havas, S, Cheung, M, Sturrock, S, Buxton, S, Cooper, A, Markowitz, S, Duran, C, et al. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Pan ZH, She ML. 1995. On karyotypes and geographical distribution of endemic genera in Umbelliferae from China. J Plant Resour Environ. 4:1–4.
- Zhong WJ, Cao L, Zhong WH, Liang J, Zhong GY. 2016. Analysis of varieties and standards of Umbelliferae medicinal plants used in Tibetan medicine. Mod Tradit Chinese Med Mater Med World Sci Technol. 18:582–588.
- Zhong GY, Wang CH, Zhou HR, Qing SY. 2008. Tibetan medicine: pharmacognosy and species consolidation. Mod Tradit Chinese Med Mater Med World Sci Technol. 10:28–32.