Abstract
The Coal Darter (Percina brevicauda) is endemic to just three watersheds within the upper Mobile River basin in Alabama. Coal Darters are imperiled and are recognized as a species of conservation priority by the Alabama Department of Wildlife and Fisheries. We have sequenced the first complete mitochondrial genome of the Coal Darter. This genomic data is currently being used to facilitate environmental DNA (eDNA) studies using Loop-mediated isothermal Amplification (LAMP) primers so that species presence and absence data can be collected within the range of the Coal Darter.
Introduction
The genus Percina currently contains 49 darter species that are only found in freshwater systems in North America (Boschung Jr and Mayden Citation2004). The Coal Darter (Percina brevicauda) has a disjunct range in the upper eastern part of the Mobile Basin above the Fall Line, where it currently occurs in the lower section of the Coosa River watershed, throughout the Cahaba River watershed, and a section of the upper Black Warrior River watershed (Suttkus et al. Citation1994; Shepard et al. Citation2004). The Coal Darter has become increasingly rare during the last 50 years, and has been recommended as threatened because of present or threatened destruction, modification, or curtailment of its habitat or range (Oronato et al. Citation2000; Warren et al. Citation2000).
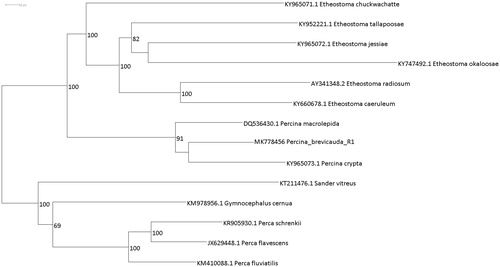
Coal Darter specimens were collected for genetic analysis at Hatchet Creek (32.916851 N, −86.270355 W; Coosa County, AL). Voucher specimens are preserved at The University of West Alabama Museum of Zoology (UWAZ 201811902). Whole genomic DNA was extracted using the Qiagen DNeasy Blood and Tissue Kit. DNA quantity and quality were assessed with a NanoDrop 2000 and gel electrophoresis, respectively. Whole genomic sequence data were generated using Illumina Hi-Seq 4000 (Illumina, CA, USA) at the Vincent J. Coates Sequencing Laboratory at The University of California, Berkeley. Paired-end reads were assembled via iterative mapping to a reference sequence with MitoBim 1.7 (Hahn et al. Citation2013) on the Cheaha cluster computer at the University of Alabama at Birmingham. The reference sequence selected for this assembly was the mitochondrial genome for the Bigscale Logperch, Percina macrolepida (GenBank DQ536430). The assembled Coal Darter mitogenome contained 16,608 bp (GenBank MK778456). Genome annotation with MitoFish 3.42 revealed that the Coal Darter mitogenome fit the typical teleost gene number and arrangement (22 tRNA’s, 13 protein coding genes, 2 ribosomal genes, and 1 control region) (Satoh et al. Citation2013). The mitogenome was checked by eye using BioEdit 7.0.5 (Hall Citation2005) and aligned to other members of the family Percidae using MAFFT (Katoh et al. Citation2017). A maximum-likelihood (ML) analysis was conducted with the GTR + G + I model in using RAxML BlackBox hosted by the CIPRES web server (www.phylo.org; Stamatakis Citation2014). The resulting topology had an ML score of -92033.181733. Bootstrap scores support a monophyletic clade of darters including the reciprocally monophyletic Percina and Etheostoma (). The Bigscale Logperch (P. macrolepida) and the Halloween Darter (Percina crypta) are in the subgenera Percina and Hadropterus, respectively. The sister taxon for P. brevicauda was not resolved with strong bootstrap support, so additional sampling among subgenera could improve this node of the tree. The publicly available mitochondrial genome sequence for the Coal Darter will aid in future development of environmental DNA (eDNA) protocols, which will contribute to attempts in identifying critical habitat and areas of occupancy for this imperiled, endemic species.
Figure 1. Interspecific phylogeny inferred under the maximum-likelihood optimality criteon (Stamatakis Citation2014). Support values are represented by bootstrap replicates in which the taxa clustered together by using the automatic halting bootstrap method (Pattengale et al. Citation2009).
Acknowledgements
Sequencing of P. brevicauda was possible with support from the Alabama Department of Conservation and Natural Resources along with support from the Birmingham Audubon Society.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boschung Jr. HT, Mayden RL. 2004. Fishes of Alabama. Washington (DC): Smithsonian Books
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Hall T. 2005. BioEdit version 7.0.0. Raleigh (NC): Department of Microbiology, North Carolina State University.
- Katoh K, Rozewicki J, Yamada KD 2017 MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief in Bioinform. [accessed date March 15, 2019], in press. doi:10.1093/bib/bbx108.
- Oronato D, Angus RA, Marion KR. 2000. Historical changes in the ichthyofaunal assemblages of the Upper Cahaba River in Alabama associated with extensive urban development in the watershed. J Freshwater Ecol. 15:47–63.
- Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A. (2009). How many bootstrap replicates are necessary? In Proceedings of the 13th Annual International Conference on Research in Computational Molecular Biology; May 18–21; Tucson. Berlin, Heidelberg: Springer. p. 184–200.
- Satoh TP, Mabuchi K, Miya M, Nishida M, Sado T, Takeshima H. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Shepard T, McGregor SW, Mettee MF, O’Neil PE. 2004. Biomonitoring in the Locust Fork Watershed, Alabama. Geol Survey of Alabama Bull. 175:1997–1998.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. 10.1093/bioinformatics/btu033
- Suttkus RD, Bart HL, Thompson BA. 1994. Two new darters, Percina (Cottogaster), from the southeastern United States: with a review of the subgenus. occasional papers/Tulane Univerity Museum of Natural History. Belle Chasse (LA): Tulane University Museum of Natural History.
- Warren ML, Burr BM, Walsh SJ, Bart HL, Cashner RC, Etnier DA, Freeman BJ, Kuhajda BR, Mayden RL, Robison HW, et al. 2000. Diversity, distribution, and conservation status of the native freshwater fishes of the southern United States. Fisheries. 25:7–31.
