Abstract
Haplosphaera phaea Handel-Mazzetti (Apiaceae) is an endangered species naturally distributed in China. The complete chloroplast genome of H. pheae was determined in this study. The chloroplast genome of H. pheae was 157,271 bp in length and divided into four distinct regions, such as large single copy region was 86,489 bp, small single copy (SSC) region was 17,892 bp, and a pair of inverted repeat regions is 26,446 bp in each length. The chloroplast genome detected a total of 130 genes including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis with the reported chloroplast genomes revealed that H. pheae is nested in the genus Hansenia.
Haplosphaera phaea Handel-Mazzetti (Apiaceae) is an herbaceous plant species naturally distributed in China. It wildly grows in forest margins and shrubs at an elevation of 3000–4200 m. Haplosphaera pheae shows unique umbel different from other species in Apiaceae which umbrella frame is sharply shortened into a spherical shape (She et al. Citation2005). Although several phylogenetic studies had reported some nuclear and chloroplast sequences of H. pheae, the complete chloroplast genome sequence is not available till now. Chloroplast genome was an important part of the whole genome. Many proteins coded from the Chloroplast gene on the genome is necessary for the process of photosynthesis. Moreover, the chloroplast genome sequence is a great molecular resource for plant taxonomy, genetic diversity, and phylogenetic system researches. At present, the complete chloroplast genome sequence of many species has been released in Apiaceae, such as Heracleum moellendorffii (Kang et al. Citation2019), Apium graveolens (Zhu et al. Citation2019), and Glehnia littoralis (Zhou et al. Citation2018). Here, we report the complete chloroplast genome sequence of H. pheae to provide a genomic resource and to clarify phylogenetic relationship of this plant with other species in the Apiaceae family.
Fresh leaves of H. pheae (accession number: J18072109-1) was collected from Mt. BaiMa (28°22′59.92″N, 99°0′18.81″E), Yunnan Province, China. The voucher specimen was stored in SZ (Sichuan University Herbarium). Total genomic DNA was extracted using Plant Genomic DNA Kit (Tiangen Biotech CO., LTD, Beijing, China).The sample of H. pheae was sequenced as the paired-end using the Illumina Novaseq 6000 platform (Illumina, Beijing, China). FastQC (v0.11.8) was used to conduct quality assessment. The filtered reads were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017) with the complete chloroplast genome of its close relative Hansenia forbesii as the reference (GenBank accession number: NC035054). The assembled chloroplast genome was annotated using Geneious 11.1.5 and corrected manually (Kearse et al. Citation2012).
The complete chloroplast genome of H. pheae (GenBank accession number: MK801097) was 157,271 bp in total sequence length with 37.60% GC contents. Four distinct regions were separated by the complete chloroplast, such as large single copy (LSC) region was 86,489 bp, small single copy (SSC) region was 17,892 bp, and a pair of inverted repeat regions are 26,446 bp in each length. The chloroplast genome detected a total of 130 genes including 85 proteincoding genes, 37 tRNA genes, and eight rRNA genes.
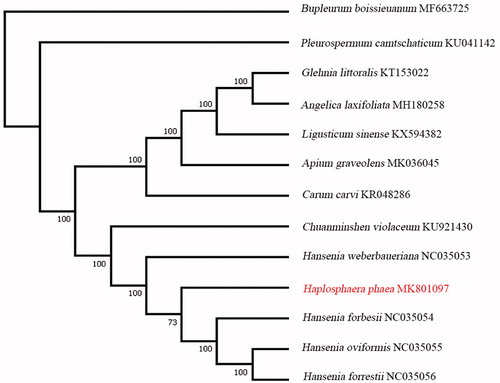
Aims at clarifying the phylogenetic relationship between H. pheae with other Apiaceae species, we generated a maximum-likelihood tree (ML) of 13 species (involving nine genus: Angelica, Apium, Bupleurum, Carum, Hansenia, Haplosphaera, Glehnia, Ligusticum, and Pleurospermum) () using RaxML (Stamatakis Citation2006) with 1000 bootstrap replicates. The complete chloroplast genome of 13 species was aligned using MAFFT (Katoh et al. Citation2002). As shown in , H. pheae was very closer to Hansenia, even nested in it, these results in accordance with early studies (Zhou et al. Citation2008, Citation2009).
Acknowledgments
The authors thank the opened cp genome data in NCBI. The authors appreciate Lu Kang, Xin Yang, Fu-min Xie and Dan-mei Su for the help of sequence analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kang L, Yu Y, Zhou SD, He XJ. 2019. Sequence and phylogenetic analysis of complete plastid genome of a medicinal plant Heracleum moellendorffii. Mitochondrial DNA B. 4:1251–1252.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- She ML, Pu FD, Pan ZH, Watson M, Cannon J, Smith H, Kljuykov E, Phillippe L, Pimenov M. 2005.Apiaceae (Umbelliferae). In: Wu ZY, Raven R, editors. Flora of China (Apiaceae through Ericaceae). Beijing: Science Press and St. Louis: Missouri Botanical Garden Press; p. 1–205.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Zhou YF, Geng ML, Li MM. 2018. The complete chloroplast genome of Glehnia littoralis, an Endangered medicinal herb of Apiaceae family. Mitochondrial DNA B. 3:1013–1014.
- Zhou J, Peng H, Downie SR, Liu ZW, Gong X. 2008. A molecular phylogeny of Chinese Apiaceae subfamily Apioideae inferred from nuclear ribosomal DNA internal transcribed spacer sequences. Taxon. 57:402–416.
- Zhou J, Gong X, Downie SR, et al. 2009. Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: Additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. MPE. 53:56–68.
- Zhu QL, Guang YL, Xiao X, Shan N, Wan CP, Yang YX. 2019. The complete chloroplast genome sequence of the Apium graveolens L. (Apiaceae). Mitochondrial DNA B. 4:463–464.