Abstract
Euphorbia tirucalli is a highly drought-tolerant shrub with a potential as biofuel source, which has an important ecological and economic value. The complete chloroplast genome sequence of E. tirucalli was carried out using Illumina sequencing platform. The complete chloroplast genome of E. tirucalli was 163,091 bp in total sequence length, which contained inverted repeats (IR) of 26,832 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 91,259 bp and 18,168 bp, respectively. The genome annotation displayed a total of 130 genes, comprising 83 protein-coding genes, 39 tRNA genes, and eight rRNA genes. The overall GC content of the plastome is 35.6%. Phylogenetic analysis with 15 species revealed that E. tirucalli was close to E. esula.
Euphorbia tirucalli L., belong to the family Euphorbiaceae, is a drought tolerant plant with a potential as biofuel source (Bernadetta et al. Citation2016). Euphorbia tirucalli has an important economic value due to rich terpenoids, sterol, and hydrocarbons in its milk, similar to petroleum component, which can be mixed directly with other substances to make biodiesel (Van Citation2001). At present, the world sped up development and utilization of new energy. The milky latex from E. tirucalli, a possible alternative to oil, was gaining widespread attention from developed countries. Biomass fuel became the newest focus of the world. In addition, E. tirucalli has a large use in folk medicine for the treatment of cancer (Melo et al. Citation2011). Therefore, we should accelerate the application and research of E. tirucalli. In plants, cpDNA provided valuable phylogenetic signals for systematic studies, owing to its conserved genome structures and comparatively high substitution rates (Wu and Ge Citation2012). In this study, we assembled and characterized the complete chloroplast genome sequence of E. tirucalli based on the lIllumina pair-end sequencing data.
Fresh leaves from an individual E. tirucalli collected in Kunming Botanical Garden (Kunming, China; 102°44′15.2″E, 25°07′04.9″N), and were used for the total genomic DNA extraction with a DNeasy Plant Mini Kit (QIAGEN, Valencia, California, USA). DNA sample and voucher specimen of E. tirucalli were deposited in the Molecular Ecology Laboratory (MEL), Institute of Loess Plateau, Shanxi University (Taiyuan, Shanxi, China). The whole-genome of E. tirucalli was sequenced using the lllumina HiseqPlatform (llumina, San Diego, CA). In total, about 500 million high-quality clean reads were obtained and used for the cp genome de nove assembly by the program NOVOPlasty (Dierckxsens et al. Citation2017) and visualized in Geneious R8 (Kearse et al. Citation2012). Annotation was performed with a command-line tool called Plann (Huang and Cronk Citation2015) and Sequin (http://www.ncbi.nlm.nih.gov/). Together with gene annotations, the complete cp genome sequences were submitted to GenBank and deposited under the accession number MH890571. A maximum-likelihood (ML) tree with 1000 bootstrap replicates was inferred using MEGA version 6 (Tamura et al. Citation2013) from alignments created by the MAFFT (Katoh and Standley Citation2013) using plastid genomes of 15 species.
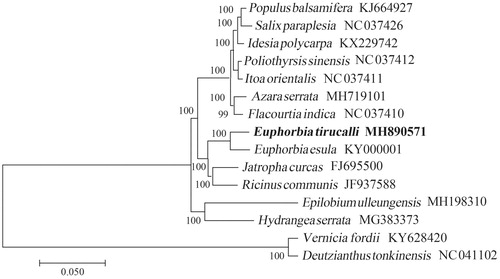
The chloroplast genome of E. tirucalli is a typical quadripartite structure with a length of 163,091 bp, which contained inverted repeats (IR) of 26,832 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 91,259 bp and 18,168 bp, respectively. The cpDNA contains 130 genes, comprising 83 protein-coding genes, 39 tRNA genes, eight rRNA genes. Among the annotated genes, five of them contain one intron (rps12, trnL-UAA, atpF, trnK-UUU, and ndhA), and five genes (ycf3, trnA-UGC, trnI-GAU, rpl2, and clpP) contain two introns. The overall GC content of the plastome is 35.6%. The phylogenetic analysis shown that E. tirucalli was closely related to the species of E. esula ().
Figure 1. The Neighbour-Joining (NJ) tree based on the 15 chloroplast genomes. The bootstrap value based on 1000 replicates is shown on each node.
This complete cp genome can be subsequently used for population, phylogenetic, and cp genetic engineering studies of E. tirucalli, and such information is vital for the intelligent use and management of this importance resources.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernadetta RH, Marina M, Filip T, et al. 2016. Euphorbia tirucalli L. – comprehensive characterization of a drought tolerant plant with a potential as biofuel source. PLoS One. 8:e63501.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Melo JG, Santos AG, Amorim ELC, Nascimento SC, Albuquerque UP. 2011. Medicinal plants used as antitumor agents in Brazil: an ethnobotanical approach. Evidbased Compl Alternat Med. 1:1–14.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30: 2725–2729.
- Van DP. 2001. Euphorbia tirucalli for high biomass production. In: Schlissel A, Pasternak D, editors. Combating desertification with plants. New York: Kluwer Academic Pub; p. 169–187.
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 62:573–578.
