Abstract
Salacia amplifolia belongs to Celastraceae. The complete plastome of S.amplifolia is 163,255 bp in length, including two inverted repeat (IR) regions of 28,932 bp, a large single-copy (LSC) region of 86,856 bp, and a small single-copy (SSC) region of 18,535 bp. The plastome contains 113 genes, consisting of 79 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of S. amplifolia is 62.50%. The complete plastome sequence of S. amplifolia will provide a useful resource for the conservation genetics of this specie as well as for the phylogenetic studies of Salacia.
Introduction
Salacia amplifolia Merr. ex Chun & F. C. How belongs to Celastraceae. It is a climbing or erect shrub, up to 4 m tall. It grows in thick forests (200–300 m) in Henan province of China (Ma et al. Citation2008). Consequently, the genetic and genomic information is urgently needed to promote its systematics research of S. amplifolia. Here, we report and characterize the complete plastome of S. amplifolia (GenBank accession number: MK799641, this study). This is the first report of a complete plastome for the genus Salacia.
In this study, S. amplifolia was sampled from Diaoluo Mountain (18.67°N, 109.88°E), which is a National Nature Reserve of Hainan, China. The voucher specimens (Wang et al. Citation2017) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
The experiment procedure is as reported in Zhu et al. (Citation2018). Around 6 Gb clean data were assembled against the plastome of Euonymus schensianus (NC_036019.1) (Wang et al. Citation2017) using MITObim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of E. schensianus (NC_036019.1). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of S. amplifolia was found to possess a total length of 163,255 bp. It has the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 28,932 bp, a large single-copy (LSC) region of 86,856 bp, a small single-copy (SSC) region of 18,535 bp. The plastome contains 112 genes, consisting of 78 unique protein-coding genes (seven of which are duplicated in the IR), 30 unique tRNA genes (seven of which are duplicated in the IR), and 4 unique rRNA genes. Among these genes, 13 genes possessed a single intron and three genes (ycf3, clpP, rps12) had two introns. The gene rps12 was found to be trans-spliced, as is typical of angiosperms. The overall A/T content in the plastome of S. amplifolia is 62.50%, which the corresponding value of the LSC, SSC, and IR region were 64.70, 68.20, and 57.20%, respectively.
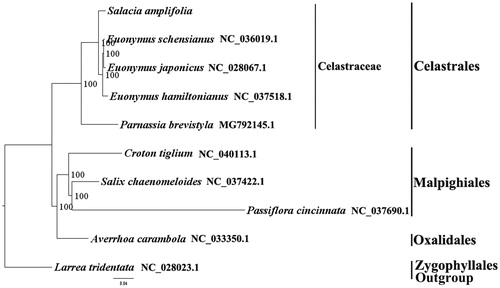
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum-likelihood (ML) phylogeny of 4 published complete plastomes of Celastrales, 3 published complete plastomes of Malpighiales and a published complete plastome of Oxalidales, using Larrea tridentata (Zygophyllaceae, Zygophyllales) as an outgroup. The phylogenetic analysis indicated that S. amplifolia is close to the genus Euonymus in this study (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of S. amplifolia will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Salacia.
Figure 1. The best maximum-likelihood (ML) phylogeny recovered from 10 complete plastome sequences by RAxML. Accession numbers – Salacia amplifolia: MK799641 (this study); Euonymus schensianus: NC_036019.1; Euonymus hamiltonianus: NC_037518.1; Euonymus japonicas: NC_028067.1; Parnassia brevistyla: MG792145.1; Croton tiglium: NC_040113.1, Salix chaenomeloides: NC_037422.1; Passiflora cincinnata: NC_037690.1; Averrhoa carambola: NC_033350.1; outgroup – Larrea tridentate: NC_028023.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Ma J-S, Zhang Z-X, Liu Q-R, Peng H, A. Michele Funston 2008. Flora of China. Beijing, China: Science Press; p. 489.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wang WC, Chen SY, Zhang XZ. 2017. Characterization of the complete chloroplast genome of the golden crane butterfly, Euonymus schensianus (Celastraceae).Conserv Genet Resour. 9:545–547.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhu Z-X, Mu W-X, Wang J-H, Zhang J-R, Zhao K-K, Ross Friedman C, Wang H-F. 2018. Complete plastome sequence of Dracaena cambodiana (Asparagaceae): a species considered “Vulnerable” in Southeast Asia. Mitochondrial DNA Part B. 3:620–621.
