Abstract
The chloroplast (cp) genome sequence of Persea americana Mill. has been characterized from Illumina pair-end sequencing. The complete cp genome was 152,732 bp in length, containing a large single copy region (LSC) of 84,632 bp and a small single copy region (SSC) of 18,346 bp, which were separated by a pair of 24,877 bp inverted repeat regions (IRs). The genome contained 126 genes, including 86 protein coding genes, 32 tRNA genes, and eight rRNA genes. Further, phylogenetic analysis of 10 representative sequences within the order Ebenales suggests that Persea americana Mill. is closely related to the species in family Sapotaceae.
Keywords:
Persea americana Mill., also known as avocado, belongs to the Lauraceae family, Ebenales order (Vergara-Pulgar et al. Citation2019), is an evergreen, heterogeneous tree species which indigenous to Mesoamerica (Boza et al. Citation2018). The fruit of avocado is a kind of climacteric fruit (Defilippi et al. Citation2018) which rich in oil content and carbohydrates, especially high in phytosterol and contain zero cholesterol, these nutrients provide a series of potential health benefits (Ervina et al. Citation2018), so it enjoys a reputation as ‘the source of life’ in Central America. Due to its nutritional properties, consumption of avocado had increased quickly worldwide in recent years (Cornelia and Christianti Citation2018; Mazhar et al. Citation2018), its planting range has also expanded to Southeast Asia countries including southern China, but its genomic data is scarce.
Herein, we reported and characterized the complete P. americana Mill. chloroplast genome. The total genomic DNA was extracted from dry leaves of P. americana Mill. plant cultivated in the plant garden of Yunnan Institute of Tropical Crops (YITC), Jinghong, China, using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina pair-end technology. The specimen of this tree and the isolated DNA were stored in YITC. The sequence reads were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017).
The total cp genome length was 152,732 bp in length, containing a large single copy region (LSC) of 84,632 bp and a small single copy region (SSC) of 18,346 bp, which were separated by a pair of 24,877 bp inverted repeat regions (IRs). The genome contained 126 genes, including 86 protein-coding genes (i.e. psbL, rpl2, rpl36, rps12, rps19, infA, ycf1, and ycf2), 32 tRNA genes (i.e. trnH-GUG, trnM-CAU, trnSI-GGA, trnD-GUC, trnG-GGC, and trnV-GAC), and 8 rRNA genes (i.e. 4.5S, 5S, 16S, and 23S rRNA). The accurate new annotated complete chloroplast genome was submitted to GenBank with accession number MK404308.
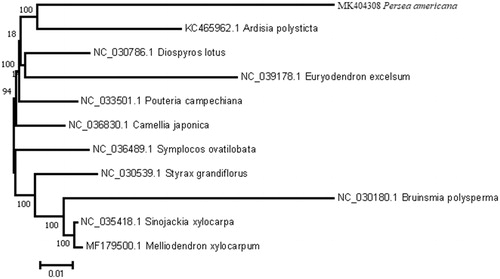
To understand the phylogenetic position of P. americana Mill. within the order Ebenales, chloroplast genome sequences of 10 other representative species in Ebenales were downloaded from the NCBI GenBank database for phylogenetic analysis. The sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and a Maximum Likelihood tree was constructed based on the other 10 sequences using MEGA7 (Kumar et al. Citation2016) with 500 bootstrap replicates. As shown in , P. americana Mill. clustered within Ebenales with strong bootstrap support.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Boza EJ, Tondo CL, Ledesma N, Campbell RJ, Bost J, Schnell RJ, Gutierrez OA. 2018. Genetic differentiation, races and interracial admixture in avocado (Persea americana Mill.), and Persea spp. evaluated using SSR markers. Genet Resour Crop Evol. 65:1195–1215.
- Cornelia M, Christianti A. 2018. Utilization of modified starch from avocado (Persea americana Mill.) seed in cream soup production. IOP Conference Series: Earth and Environmental Science.
- Defilippi BG, Ejsmentewicz T, Covarrubias MP, Gudenschwager O, Campos-Vargas R. 2018. Changes in cell wall pectins and their relation to postharvest mesocarp softening of “Hass” avocados (Persea americana Mill.). Plant Physiol Biochem. 128:142–151.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Ervina Surjawan I, Abdillah E. 2018. The potential of avocado paste (Persea americana) as fat substitute in non-dairy ice cream. IOP Conference Series: Earth & Environmental Science.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Mazhar M, Joyce D, Hofman P, Vu N. 2018. Factors contributing to increased bruise expression in avocado (Persea americana M.) cv. ‘Hass’ fruit. Postharvest Biol Technol. 143:58–67.
- Vergara-Pulgar C, Rothkegel K, González-Agüero M, Pedreschi R, Meneses C. 2019. De novo assembly of Persea americana cv. ‘Hass’ transcriptome during fruit development. BMC Genomics. 20:108. doi:10.1186/s12864-019-5486-7.