Abstract
Microdous chalmersi is an Odontobutidae fish that mainly inhabits in East and Southeast Asia. In this study, the complete mitochondrial genome of M. chalmersi was first determined and described. It was 16,485 bp in length and composed by 13 protein-coding genes, 22 tRNA genes, two rRNA genes, a putative light strand origin of replication (OL) and a putative control region (D-loop). The overall nucleotide composition was 28.9% of A, 16.0% of G, 25.0% of T, and 30.1% of C, with a total of 53.9% A + T content. Phylogenetic analysis suggested that M. chalmersi had the closest evolutionary relationship with Micropercops swinhonis. The availability of the M. chalmersi mitogenome would contribute to further phylogenetic research of Odontobutidae and genetics studies of this species.
Keywords:
Microdous chalmersi, formerly known as Philypnus chalmersi, is a small ornamental Odontobutidae fish with vomerine teeth, which mainly distributes in East and Southeast Asia (Nichols and Pope Citation1927; Chen et al. Citation2002). The taxonomic status of M. chalmersi was confirmed based on new morphological evidence and multiple nuclear loci until recently (Li et al. Citation2018), but no complete mitogenome has been reported. In this study, we provided the first characterization of the complete mitochondrial genome of M. chalmersi, contributing to molecular and phylogenetic studies of this fish species.
The sample of M. chalmersi was collected from Dahua Yao Autonomous County (23°33′35.36″N, 107°20′36.06″E) in the upper Pearl River drainage basin (Guangxi, China). Muscle specimens were fixed in 95% ethanol and preserved in Guangxi Colleges and Universities Key Laboratory of Aquatic Healthy Breeding and Nutrition Regulation, Guangxi University. Total genomic DNA was extracted through Genomic DNA Isolation Kit (QIAGEN, Hilden, Germany). The sequence was amplified through PCR with ten primer pairs and then annotated using MitoAnnotator (Iwasaki et al. Citation2013).
The complete mitogenome of M. chalmersi was 16,485 bp in length (GenBank accession no. MH644035) and was A + T-biased (53.9%) with a base composition of 28.9% A, 25.0% T, 16.0% G, and 30.1% C. Genomic organization showed high degree of conservation among M. chalmersi and other Odontobutidae fishes, including 13 PCGs, 22 tRNA genes, 2 rRNA genes, a putative origin of light-strand replication and an AT-rich control region (D-loop). Twelve PCGs initiated with ATG while COI shared GTG as start codon. Six PCGs had the common TAA termination codon, whereas the remaining seven were terminated by incomplete stop codon T–– or TA–. Two rRNAs (949 bp 12S and 1679 bp 16S) were composed of 52.2% AT, and the D-loop was 849 bp long with the AT content of 64.7%.
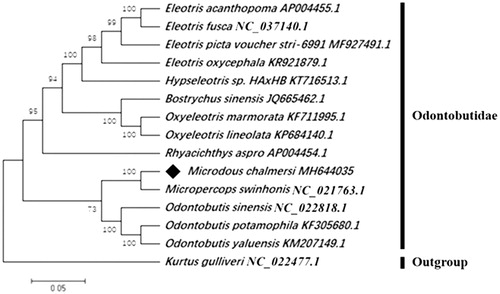
MEGA 7.0 software was used for constructing a maximum-likelihood (ML) tree based on the concatenated supergene consisting of 13 protein-coding genes (PCGs) (Kumar et al. Citation2016). Evolutionary model selection was performed for the concatenated alignment using PhyML 3.0 software, where the GTR + G + I was identified as the most appropriate to the data (Guindon et al. Citation2010). Molecular phylogenetic analysis revealed that M. chalmersi had the closest evolutionary relationship with Micropercops swinhonis (), which was similar to recent morphological classification and molecular works (Li et al. Citation2018). The present study of M. chalmersi preliminary revealed its mitochondrial genetic background and will provide essential information for further genetic investigations.
Figure 1. Phylogenetic tree of M. chalmersi, 13 related Odontobutidae fishes and one outgroup from kurtidae based on 13 concatenated mitochondrial protein coding genes by Maximum Likelihood (ML) analysis with GTR + G + I model. The number at each node is the bootstrap probability. The number after the species name is the GenBank accession number. The genome sequence of M. chalmersi is marked with rhombus.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for this paper.
Additional information
Funding
References
- Chen IS, Kottelat M, Wu HL. 2002. A new genus of freshwater sleeper (Teleostei: Odontobutididae) from southern China and Mainland Southeast Asia. J Fish Soc Taiwan. 29:229–235.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. Mitofish and Mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li HJ, He Y, Jiang JM, Liu ZZ, Li CH. 2018. Molecular systematics and phylogenetic analysis of the Asian endemic freshwater sleepers (Gobiiformes: Odontobutidae). Mol Phylogenet Evol. 121:1–11.
- Nichols JT, Pope CH. 1927. The fishes of Hainan. B Am Mus Nat Hist. 54:321–394.
