Abstract
The Eurasian or Greek blindsnake, Xerotyphlops vermicularis, is the only European representative of the infraorder Scolecophidia, a largely understudied group of snakes with a poor record of available mitogenomes. Its complete mitochondrial genome, the first reported for the genus Xerotyphlops, was assembled through next-generation sequencing. Its total length is 16,568 bp and includes 22 tRNAs, 2 ribosomal RNA genes, 13 protein-coding genes, and a control region, showing the typical gene-arrangement for Typhlopidae. Eight tRNAs and ND6 are encoded on the light strand, while all other genes are encoded on the heavy strand.
Snake are divided into two major infraorders, the ‘true snakes’ Alethinophidia and the ‘wormsnakes’ Scolecophidia. The latter are small to medium-sized burrowing snakes with no external eyes that, despite their worldwide distribution (except Antarctica) and their great number of species, they have been very poorly studied (Nagy et al. Citation2015). Their higher-level phylogenetics, systematics, and taxonomy have only recently been assessed (Adalsteinsson et al. Citation2009; Vidal et al. Citation2010; Kornilios et al. Citation2013; Hedges et al. Citation2014; Pyron and Wallach Citation2014; Nagy et al. Citation2015; Miralles et al. Citation2018). They include more than 400 species and about 40 genera organized in five families: Anomalepididae, Gerrhopilidae, Typhlopidae, Leptotyphlopidae and Xenotyphlopidae (Uetz et al. Citation2019).
The available mitochondrial genomes for Scolecophidia are very few. In fact, only five mitogenomes have been sequenced, representing five species and genera and three of the families. Here I present the complete mitochondrial genome of Xerotyphlops vermicularis Merrem, 1820, which is also the first sequenced mitogenome of this genus, as a new addition to the poor existing record of wormsnake mitogenomes. Xerotyphlops vermicularis, known as the Eurasian or Greek blindsnake, belongs to Typhlopidae and is the only extant scolecophidian representative occurring in Europe. It is most probably a species-complex (Kornilios et al. Citation2011, Citation2012; Kornilios Citation2017) and its type locality is ‘Greece’. An adult specimen was collected in 2009 in Makri (Thrace, Greece; 40.853932°, 25.747042°) and deposited in the Biology Department, University of Patras (sample code TV071). Total genomic DNA was extracted and used, together with other squamate samples, in a comparative analysis that utilized ultraconserved elements (UCEs) under a sequence capture protocol (Faircloth et al. Citation2012). The 150 bp paired-end library was prepared using the Truseq Nano DNA kit (Illumina Inc., San Diego, CA) and sequenced on a single Illumina HiSeq 4000 lane. The mitogenome was assembled de novo from the off-target sequences with NOVOPlasty 2.7.1 (Dierckxsens et al. Citation2017). Genes were annotated using the MITOS WebServer (Bernt et al. Citation2013) and checked manually. Alignment with all available scolecophidian mitogenomes was done with MAFFT (Katoh et al. Citation2017). This dataset, after removing the poorly aligned control region, was used to reconstruct a maximum-likelihood (ML) tree with IQ-TREE1.4.3 (Nguyen et al. Citation2015), using 1000 ultrafast bootstrap alignments (Minh et al. Citation2013).
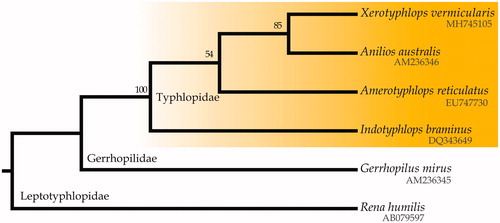
The Eurasian blindsnake’s mitogenome (Genbank accession number MH745105) is 16,568 bp long and includes 22 transfer RNA genes (duplicate tRNA-Leu and tRNA-Ser), 2 ribosomal RNA genes, 13 protein-coding genes, and a control region, showing the typical gene-arrangement for Typhlopidae (type II in Qian et al. Citation2018). Eight tRNAs and ND6 are encoded on the light strand, while all others are encoded on the heavy strand. The overall composition of the heavy strand is A (33.0%), T (27.7%), C (26.2%), and G (13.1%). The ML mitogenomic phylogeny, rooted with the leptotyphlopid Rena humilis, is shown in . Within the monophyletic unit of Typhlopidae, Amerotyphlops represents the South America group of typhlopids, while Xerotyphlops, Indotyphlops, and Anilios represent the Asian one, as resulted in all contemporary DNA phylogenies (e.g. Miralles et al. Citation2018). The presented tree does not exhibit this phylogenetic configuration, but it is the very limited available scolecophidian mitogenomes and the overall representation gap that is responsible for this conflict.
Acknowledgements
The sequence-capture work took place in the Leaché Lab (Department of Biology, University of Washington). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant.
Disclosure statement
The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
Additional information
Funding
References
- Adalsteinsson SA, Branch WR, Trape S, Vitt LJ, Hedges SB. 2009. Molecular phylogeny, classification, and biogeography of snakes of the family Leptotyphlopidae (Reptilia, Squamata). Zootaxa. 2244:1–50.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst Biol. 61:717–726.
- Hedges SB, Marion AB, Lipp KM, Marin J, Vidal N. 2014. A taxonomic framework for typhlopid snakes from the Caribbean and other regions (Reptilia, Squamata). Caribb Herpet. 49:1–61.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. bbx108.
- Kornilios P. 2017. Polytomies, signal and noise: revisiting the mitochondrial phylogeny and phylogeography of the Eurasian blindsnake species complex (Typhlopidae, Squamata). Zool Scr. 46:665–674.
- Kornilios P, Giokas S, Lymberakis P, Sindaco R. 2013. Phylogenetic position, origin and biogeography of Palearctic and Socotran blind-snakes (Serpentes: Typhlopidae). Mol Phylogenet Evol. 68:35–41.
- Kornilios P, Ilgaz Ç, Kumlutaş Y, Giokas S, Fraguedakis-Tsolis S, Chondropoulos B. 2011. The role of Anatolian refugia in herpetofaunal diversity: an mtDNA analysis of Typhlops vermicularis Merrem, 1820 (Squamata, Typhlopidae). Amphibia-Reptilia. 32:351–363.
- Kornilios P, Ilgaz Ç, Kumlutaş Y, Lymberakis P, Moravec J, Sindaco R, Rastegar-Pouyani N, Afroosheh M, Giokas S, Fraguedakis-Tsolis S, et al. 2012. Neogene climatic oscillations shape the biogeography and evolutionary history of the Eurasian blindsnake. Mol Phylogenet Evol. 62:856–873.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30:1188–1195.
- Miralles A, Marin J, Markus D, Herrel A, Hedges SB, Vidal N. 2018. Molecular evidence for the paraphyly of Scolecophidia and its evolutionary implications. J Evol Biol. 31:1782–1793.
- Nagy ZT, Marion AB, Glaw F, Miralles A, Nopper J, Vences M, Hedges SB. 2015. Molecular systematics and undescribed diversity of Madagascan scolecophidian snakes (Squamata: Serpentes). Zootaxa. 4040:31–47.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Pyron RA, Wallach V. 2014. Systematics of the blindsnakes (Serpentes: Scolecophidia: Typhlopoidea) based on molecular and morphological evidence. Zootaxa. 3829:1–81.
- Qian L, Wang H, Yan J, Pan T, Jiang S, Rao D, Zhang B. 2018. Multiple independent structural dynamic events in the evolution of snake mitochondrial genomes. BMC Genomics. 1019:354.
- Uetz P, Freed P, Hošek J. 2019. The Reptile Database; [accessed 2019 April 20]. http://www.reptile-database.org.
- Vidal N, Marin J, Morini M, Donnellan S, Branch WR, Thomas R, Vences M, Wynn A, Cruaud C, Hedges SB. 2010. Blindsnake evolutionary tree reveals long history on Gondwana. Biol Lett. 6:558–561.