Abstract
The complete mitogenome of Argiope perforata (GenBank accession number MK512574) is 14,032 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes, and an A + T-rich region. ATT, ATA, TTA, TTG were initiation codons and TAA, TAG, and T were termination codons. Ten tRNAs (trnD, trnW, trnL2, trnE, trnF, trnR, trnH, trnP, trnT, and trnL1) lacked the TΨC arm stem, while three tRNAs (trnA, trnS1, and trnS2) lost the dihydrouracil (DHU) arm. Phylogenetic tree based on 13 PCGs showed that A. perforata is closely related to Neoscona theisi, and clustered within Araneidae clade.
Keywords:
The orb-weaving spider Argiope perforata belongs to the genus of Argiope, which includes 81 known species around the world (Wang Citation1988; Platnick Citation2015). Most of these spiders often have a strikingly coloured abdomen and are important predators of insect pests (Levi Citation2004). In this study, adult species of A. perforata were collected from Maolan Nature Reserve in Libo country, Guizhou Province, China (N25°18′, E107°52′), and deposited in the spider specimen room of Guiyang University with an accession number GYU-GZML-13.
The complete mitogenome of A. perforata (GenBank accession number MK512574) is 14,032 bp in length, and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rrnL and rrnS), and an A + T-rich region. The gene order and orientation of A. perforata are consistent with those of other known spider mitogenomes (Li et al. Citation2016; Xu et al. Citation2019). Fifteen genes were transcribed on the minor strand (J-strand), while the others were encoded on the major strand (N-strand). The overall base composition of A. perforata mitogenome is 35.30% for A, 9.65% for C, 38.90% for T, and 16.15% for G, with a high AT bias of 74.20%. The AT-skew and GC-skew of this genome were −0.049 and 0.252, respectively.
A total of 22 bp overlaps have been found at five gene junctions of A. perforata mitogenome, and the length of overlaps are ranging from 3 to 7 bp. There are 25 intergenic spacer sequences in a total of 208 bp with length varying from 1 to 26 bp and the largest intergenic spacer is located between nad3 and trnL2. The length of 22 tRNAs ranged from 52 bp (trnA) to 65 bp (trnM), A + T content ranged from 68.52% (trnR) to 83.61% (trnC). Thirteen tRNAs lack the potential to form the cloverleaf secondary structure. Ten of them (trnW, trnD, trnL2, trnE, trnF, trnR, trnH, trnP, trnT, and trnL1) lacked the TΨC arm stem, three tRNAs (trnA, trnS1, and trnS2) lost the dihydrouracil (DHU) arm. The rrnL and rrnS genes are 1022 and 695 bp in length, with the A + T contents of 78.47 and 75.97%, respectively. The A + T-rich region of this mitogenome is 451 bp in length with an A + T content of 79.16% and located between trnQ and trnM.
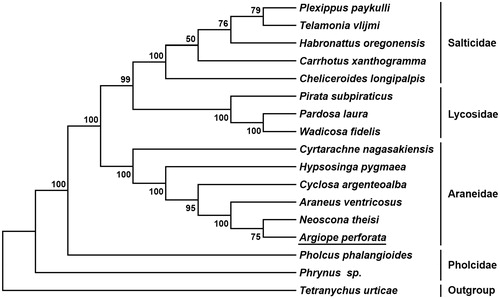
Nine PCGs start with a typical ATN (ATT and ATA) codon, three genes (cox2, cox3, and nad6) start with TTG, and cox1 uses TTA as initiation codon. Twelve PCGs terminate with conventional stop codons (TAA and TAG), while nad4 uses incomplete codon (T) as termination codon. Based on the concatenated amino acid sequences of 13 PCGs, the neighbor-joining method was used to construct the phylogenetic relationship of A. perforata with 15 other spiders (Kumar et al. Citation2016). The result showed that A. perforata is closely related to Neoscona theisi, and clustered within Araneidae clade ().
Figure 1. Phylogenetic tree showing the relationship between Argiope perforate and 15 other spiders based on neighbor-joining method. GenBank accession numbers used in the study are the following: Araneus ventricosus (KM588668), Carrhotus xanthogramma (NC_027492.1), Cheliceroides longipalpis (NC_041120.1), Cyclosa argenteoalba (KP862583), Cyrtarachne nagasakiensis (KR259802), Habronattus oregonensis (AY571145), Hypsosinga pygmaea (KR259803), Neoscona theisi (NC_026290.1), Pardosa laura (NC_025223.1), Pholcus phalangioides (NC_020324.1), Phrynus sp. (NC_010775.1), Pirata subpiraticus (NC_025523.1), Plexippus paykulli (NC_024877.1), Telamonia vlijmi (NC_024287.1), Tetrancychus urticae (EU345430.1), and Wadicosa fidelis (NC_026123.1). T. urticae was used as an outgroup. The spider determined in this study is underlined.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Levi HW. 2004. Comments and new records for the American genera Gea and Argiope with the description of a new species (Araneae: Araneidae). Bull Mus Comp Zool. 158:47–66.
- Li C, Wang ZL, Fang WY, Yu XP. 2016. The complete mitochondrial genomes of two orb-weaving spider Cyrtarachne nagasakiensis (Strand, 1918) and Hypsosinga pygmaea (Sundevall, 1831) (Araneae: Araneidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2811–2812.
- Platnick NI. 2015. The world spider catalog, version 13.5. American museum of natural history. http://research.amnh.org/iz/spides/catalog/.
- Wang L. 1988. Description of the male of Argiope perforata (Araneae: Araneidae). Acta Zootaxon Sin. 13:101–102.
- Xu KK, Lin XC, Yang DX, Yang WJ, Li C. 2019. Characterization of the complete mitochondrial genome sequence of Neoscona scylla and phylogenetic analysis. Mitochondrial DNA Part B. 4:416–417.
