Abstract
Trinket cattle were introduced to Trinket Island of Nicobar archipelago by Danish people during their early colonization period. These cattle became feral in nature after Great Sumatra earthquake and Indian Ocean Tsunami since 26 December 2004. Negligence has brought the cattle on the brink of extinction. In the present study, we document the complete mitochondrial genome sequence of Trinket Cattle. The mitogenome contains 37 genes including 13 protein-coding genes, 22 tRNAs, and two ribosomal RNA genes. In addition, a control region (D-loop) was also present. Phylogenetic analysis showed that Trinket Cattle belongs to Bos indicus group. The results of the study will be helpful for formulating of conservation strategy of the highly endangered breed.
Keywords:
Trinket cattle, a highly endangered group of cattle are available in Trinket island of Nicobar group of Islands. Early historical documents suggest that Danish people introduced these cattle in Trinket Island during their initial phases of colonization (Kloss Citation1903). When the Danish colonization came to an end, the cattle were looked after by indigenous tribal people. The island was left abandoned by the aboriginal tribe after Great Sumatra earthquake and Indian Ocean Tsunami since 26 December 2004. Therefore, the herd of cattle became totally feral in nature. Negligence has brought the cattle to the brink of extinction; around 150 of the descendants of cattle are now available. This is extremely important to characterize the animals and take proper conservation measures to save them from extinction. In the present study, the whole mitochondrial DNA of Trinket cattle was sequenced and characterized.
Blood sample was collected from Trinket cattle (Trinket Island, Nicobar Group of Islands, 8.08°N 93.58°E). Isolated DNA sample is preserved in the repository of ICAR-CIARI, Port Blair. Enrichment of mitochondrial DNA was done by following the protocol of NEBNext Microbiome Enrichment Kit (New England Biolab Inc). Library preparation was done by using Nextera XT DNA Library Preparation Kit, Illumina. The library was sequenced on Illimina Nextseq platform with paired-end sequencing. After sequencing, the filtered reads were assembled using a reference sequence, Bos indicus (Zwergzebu breed, Genbank accession no. AF492350) complete mitochondrion.
The complete mitochondrial DNA sequence of Trinket cattle was deposited to GenBank with accession number MK335920. The mitogenome was 16,341 bp in length and consisted of 37 genes including 13 protein-coding genes (PCGs), 22 tRNAs, and two rRNAs. One A + T rich region (D-loop) of 913 bp was also present. In the mitogenome, a total of 12 overlapping regions encompassing 152 bp was observed. In addition, 13 intergenic spacer regions ranging from 1 bp to 4 bp were found.
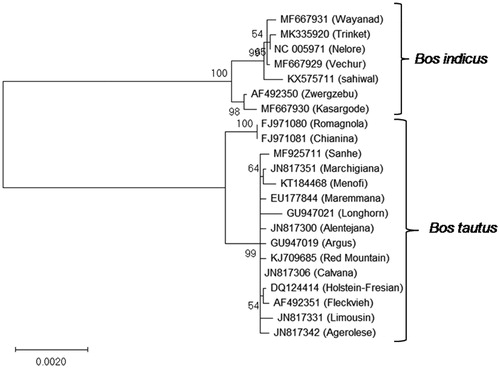
Cattle belong to the family Bovidae and two main species of domestic cattle taurine (Bos tautus) and zebu (Bos indicus) have been reported. Archaeological and genomic evidence suggests that zebu cattle was originated in India and later spread to Africa and southeast Asia (Naik Citation1978). On the contrary, the most likely domestication of the taurine breeds happened in Fertile Crescent (Beja-Pereira et al. Citation2006). Whole mitochondrial sequences of zebu and taurine breeds were retrieved from Genbank. Phylogenetic relationship was analysed based on concatenated sequence of protein-coding genes. Phylogenetic analysis was done by maximum-likelihood (ML) method with 1000 bootstrap replications using MEGA-X (Kumar et al. Citation2018). From the phylogenetic tree (), it was found that Bos indicus and Bos taurus formed two separate clads and Trinket cattle was within in the Bos indicus clad indicating that Trinket cattle was introduced from Indian subcontinent by Danish people.
Figure 1. Phylogenetic analysis of Trinket cattle based on concatenated sequence of PCGs. Phylogenetic relationship between mtDNA sequences of Trinket cattle and other mitogenomes of cattle were analyzed using maximum-likelihood method based on the Hasegawa–Kishino–Yano model (Hasegawa et al. Citation1985). Accession numbers of cattle breeds used are as follows: MK335920 (Trinket), AF492350 (Zwergzebu), KX575711 (Sahiwal), MF667929 (Vechur), MF667930 (Kasargode), MF667931 (Wayanad), NC_005971 (Nelore), AF492351 (Fleckvieh), DQ124414 (Holstein-Fresian), EU177844 (Maremmana), FJ971080 (Romagnola), FJ971081 (Chianina), GU947019 (Argus), GU947021 (Longhorn), JN817300 (Alentejana), JN817306 (Calvana), JN817331 (Limousin), JN817342 (Agerolese), JN817351 (Marchigiana), KJ709685 (Red Mountain), KT184468 (Menofi), and MF925711 (Sanhe).
The current study presents the mitogenome analysis of Trinket Cattle; the results of the study will be helpful to understand the genetic root of the breed and for sketching of conservation strategy of the highly endangered breed.
Acknowledgements
Authors are very thankful to the Chairperson, Kamorta Tribal Council, Nicobar for her help in collection of biological samples from trinket Cattle.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Beja-Pereira A, Caramelli D, Lalueza-Fox C, Vernesi C, Ferrand N, Casoli A, Goyache F, Royo LJ, Conti S, Lari M, et al. 2006. The origin of European cattle: evidence from modern and ancient DNA. Proc Natl Acad Sci USA. 103:8113–8118.
- Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22:160–174.
- Kloss BC. 1903. In the Andamans and Nicobars. London: John Murray.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Naik SN. 1978. Origin and domestication of Zebu cattle (Bos indicus). J Hum Evol. 7:23–30.
