Abstract
Saltator similis, popularly known as Green-winged Saltator, is one of the most trafficked species in Brazil. DNA from a muscle tissue sample was sequenced on MiSeq (Illumina) sequencer. The reads were assembled to reference using Geneious. The mtDNA consisted of 16,750 base pairs containing 2 ribosomal RNA, 22 transporter RNA, 13 protein-coding genes, and 1 control region. Most of the tRNA and PCGs were encoded on the heavy strand.
Saltator similis (d'Orbigny & Lafresnaye, 1837 in Guala & Döring Citation2019) popularly known as Green-winged Saltator, is a member of order Passeriformes. It inhabits in parts of Brazil, Bolivia, Uruguay, Paraguay and Argentina (BirdLife International Citation2018). The current decrease in population is due to the loss of habitats and the illegal capture (UFV Citation2009, Manhães & Loures-Ribeiro Citation2011). Saltator similis is one of the most seized species in actions to combat the illegal trade of wild animals in Brazil (Borges et al. Citation2006; SEMA/PMA Citation2006; Gogliath et al. Citation2010; Destro Citation2012; Nunes et al. Citation2012; Franco et al. Citation2012; Felker et al. Citation2013; Zocche and Vianna Citation2013; Silva Citation2015; Freitas et al. Citation2015). In Brazilian legislation, captive breeding is permitted. To be a legal bird, it must be born in captivity and have an official inviolable band.
A population of female individual from Saltator similis species deceased as a consequence of illegal trafficking. It was identified and donated by IBAMA (Brazilian Environmental Agency) with proper licensing from the competent authorities (SISBIO 56471-1, IEF 024/2016 and CEUA-UFMG 37/2017) and mitochondrial DNA was extracted following the protocol from Françoso et al. (Citation2015). The specimen was taxidermized and is in the Center of Taxonomic Collections of Federal University of Minas Gerais (voucher DZ7289).
DNA library was constructed and sequenced using a single-read 300-bp strategy on a MiSeq system (Illumina, San Diego, CA) using MiSeq Reagent Kit V2-300. Reads were trimmed with Phred 30 and assembled to reference (mtDNA of Geospiza magnirostris – GenBank MG682351.1) using Geneious 11.1.5 (https://www.geneious.com). The complete mitochondrial DNA of S. similis (GenBank accession number MK419316) was found to be circular in shape with 16,750 bp length, with average coverage of 148 reads. mtDNA was submitted to nucleotide BLAST (Altschul et al. Citation1990), which resulted in high similarity with the mitogenome of other Passeriformes species. The mtDNA displayed a GC content of 46.93% and base frequencies were 30% A, 14.36% G, 23% T, and 32.57% C.
The mitogenome was annotated with MITOS (Bernt et al. Citation2013) and verified with ExPASy (Gasteiger et al. Citation2003). Genes in S. similis were arranged similarly to other avian mitogenomes (Raposo do Amaral et al. Citation2015; Ludwig et al. Citation2017; Liang et al. Citation2018), with a different organization of the genes near the control region when compared to typical vertebrate mitogenome (Quinn and Wilson Citation1993). The mtDNA structure contained 2 rRNA, 22 tRNA, 13 protein-coding genes (PCGs), and 1 control region (D-loop). Protein-coding genes commonly had ATG as start codon (11 PCGs). Nine of the 13 PCGs contained a TAA stop codon. Eight of the 22 tRNAs and one PCG (Nd6) were encoded on the light (L) strand. The remaining genes were encoded on the heavy (H) strand. We found 10 overlapping regions.
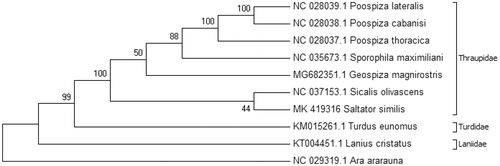
Phylogenetic analyses were conducted using MEGA version 6.06 (MEGA Inc., Englewood, NJ) (Tamura et al. Citation2013) using the neighbor-joining method (Saitou and Nei Citation1987). D-loop region was excluded from phylogenetic analysis due to its hypervariability (Gonder et al. Citation2007) ().
Figure 1. Consensus neighbor-joining tree of 10 avian species. Analyses were conducted in MEGA6 (Tamura et al. Citation2013). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein Citation1985). The evolutionary distances were computed using the Kimura 2-parameter method (Kimura Citation1980) and are in the units of the number of base substitutions per site. The analysis involved 10 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd. Thraupidae Family grouped together with a high boostrap value including Saltator similis. Turdidae and Laniidae Family were grouped in separate clades. Tree was rooted using Ara ararauna as the outgroup.
Acknowledgments
We thank IBAMA, IEF and colleagues of the Laboratory of Biotechnology and Molecular Markers for their partnership. We thank Jean Oliveira and team of Taxonomy Collection Center for their collaborations.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- BirdLife International. 2018. Species factsheet: Saltator similis; [accessed 2018 Oct 25]. http://www.birdlife.org.
- Borges RC, Oliveira A, Bernardo N, Costa RMMC. 2006. Diagnóstico da fauna silvestre apreendida e recolhida pela Polícia Militar de Meio Ambiente de Juiz de Fora, MG (1998 e 1999). Revista Brasileira de Zoociências. 8:23–33.
- Destro GFG. 2012. Efforts to combat wild animals trafficking in Brazil. In: Lammed GA, editor. Biodiversity enrichment in a diverse world. Sao Paolo: InTech; p. 421–436.
- Felker RM, Dörr AC, Rovedder AP, Piazza EM, Dick G. 2013. Levantamento parcial da avifauna apreendida pelo Escritório Regional do Ibama de Santa Maria-RS. Revista Eletrônica em Gestão, Educação e Tecnologia Ambiental, Santa Maria, v 11, n. 11, p. 2506–2510; [accessed 2016 Aug 18]. http://cascavel.ufsm.br/revistas/ojs-2.2.2/index.php/reget/article/download/8734/pdf
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783–791.
- Franco MR, Câmara FM, Rocha DCC, Souza RM, Oliveira NJF. 2012. Animais silvestre apreendidos no período de 2002 a 2007 na macrorregião de Montes Claros. Minas Gerais. Enciclopédia Biosfera, Centro Científico Conhecer. 8:1007–1018.
- Françoso E, Gomes F, Arias MC. 2015. A protocol for isolating insect mitochondrial genomes: a case study of NUMT in Melipona flavolineata (Hymenoptera: Apidae). Mitochondrial DNA Part A. 27:1–4.
- Freitas ACP, Oviedo-Pastrana ME, Vilela DAR, Pereira PLL, Loureiro LOC, Haddad JPA, Martins NRS, Soares DFM. 2015. Diagnóstico de animais ilegais recebidos no centro de triagem de animais silvestres de Belo Horizonte, Estado de Minas Gerais, no ano de 2011. Cienc Rural. 45:163–170.
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784–3788.
- Gogliath M, Bisaggio EL, Ribeiro LB, Resgalla AE, Borges RC. 2010. Avifauna apreendida e entregue voluntariamente ao Centro de Triagem de Animais Silvestres (Cetas) do Ibama de Juiz de Fora, Minas Gerais. Atualidades Ornitológicas on-Line; 154:55–59 [accessed 2016 Aug 18]. http://www.ao.com.br/download/ao154_55.pdf.
- Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. 2007. Whole-mtDNA genome sequence analysis of ancient African Lineages. Mol Biol Evol. 24:757–768.
- Guala G, Döring M. 2019. Integrated Taxonomic Information System (ITIS). National Museum of Natural History, Smithsonian Institution. [accessed via GBIF.org on 2019-05-17] Checklist dataset https://doi.org/10.15468/rjarmt
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Liang W, Yunlin Z, Zhenggang X, Tian H, Libo Z, Shiquan L. 2018. The complete mitochondrial genome and phylogeny of Geospiza magnirostris (Passeriformes: Thraupidae). Conserv Gen Res. 2:1–3.
- Ludwig S, Martins APV, Queiroz ALL, Carmo AO, Oliveira-Mendes BBR, Kalapothakis E. 2017. Complete mitochondrial genome of Sporophila maximiliani (Ave, Passeriformes). Mitochondrial DNA Part B. 2:2:417–418.
- Manhães MA, Loures-Ribeiro A. 2011. The avifauna of the Poço D’Anta Municipal Biological Reserve, Juiz de Fora, MG. Biota Neotrop. 11:275–286.
- Nunes PB, Barreto AS, Franco EZ. 2012. Subsídios à ação fiscalizatória no combate ao tráfico de aves silvestres e exóticas em Santa Catarina. Ornithologia. Matinhos, n. 5, n. 1, p. 28–33; [accessed 2016 Aug 2]. http://www.cemave.net/publicacoes/index.php/ornithologia/article/view/75/78.
- Quinn TW, Wilson AC. 1993. Sequence evolution in and around the mitochondrial control region in birds. J Mol Evol. 37:417–425.
- Raposo do Amaral F, Neves LG, Resende MF Jr, Mobili F, Miyaki CY, Pellegrino KC, Biondo C. 2015. Ultraconserved elements sequencing as a low-cost source of complete mitochondrial genomes and microsatellite markers in non-model amniotes. PLoS One. 10:e0138446.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- SEMA/PMA. Polícia Militar Ambiental De São Paulo (PMA-SP). 2006. Tráfico de Animais Silvestres da Fauna Nacional – Dados estatísticos e estratégias operacionais 2001–2005; [accessed 2016 Aug 3]. http://www.pea.org.br/educativo/relatorio_policia_ambiental.pdf
- Silva NS. 2015. Espécimes recebidos no centro de triagem de animais silvestres de salvador/BA durante os anos de 2012 a 2014. Trabalho de Conclusão de Curso. Salvador: Universidade Federal da.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Universidade Federal De Viçosa (UFV). 2009. Museu de Zoologia João Moojen. Bicho da vez n° 4. Trinca-ferro-verdadeiro (Saltator similis); [accessed 2018 Dec 15]. http://www.museudezoologia.ufv.br/bichodavez/edicao04.pdf
- Zocche JJ, Viana IR. 2013. Avifauna apreendida no extremo sul catarinense: apreensões feitas durante oito anos de fiscalização e combate à captura de aves silvestres. Revista Brasileira de Biociências. Porto Alegre. 11:395–404.
