Abstract
Acer catalpifolium is a rare and endangered species in southwestern China. In this study, the complete chloroplast genome sequence of A. catalpifolium was characterized by de novo assembly using high-throughput sequencing. The length of the whole chloroplast genome was 156,225 bp, containing a large single copy region (LSC) of 85,976 bp and a small single copy region (SSC) of 18,066 bp, which were separated by a pair of 26,092 bp inverted repeat regions (IRs). The sequence contained 135 unique genes, including 40 tRNA, eight rRNA, and 87 protein-coding genes. The overall GC content of the chloroplast genome is 37.9% and those in the LSC, SSC, and IR regions are 36.1%, 32.2%, and 42.9%, respectively. The phylogenetic analysis based on reported chloroplast sequences of Sapindaceae showed that A. catalpifolium is sister to A. miaotaiense, and the genera of Acer and Dipteronia are closely related to Aesculus.
Acer is a genus of woody plants in subfamily Hippocastanoideae of Sapindaceae (formerly in Aceraceae) (Harrington et al. Citation2005; APG Citation2016). Acer catalpifolium Rehder, a tall deciduous tree, is endemic to Sichuan and Guizhou provinces, southwestern China, scattering in subtropical evergreen broad-leaved forests or mixed forests at the altitudes of 400–1100 m (Wu et al. Citation2018; Zhang et al. Citation2018). It has been classified as a rare and endangered plant in China (Fu Citation1992), and was recently listed as one of the 33 plant species with extremely small populations (PSESP) (Ma et al. Citation2013) in Sichuan Province (Pan et al. Citation2014). Based on our field investigations for PSESP in 2017 and 2018, the wild population of A. catalpifolium now is minimal and needs urgent conservation. In this study, we assembled the complete chloroplast genome of A. catalpifolium using next-generation sequencing to provide a gene source for further genetic and conservation studies.
Fresh young leaves of A. catalpifolium were collected from Qingcheng Mountain in Sichuan Province, China (N 30°53′35.27″, E103°32′42.25″), and its voucher specimen (with collection number of BFUZLC013-001) was deposited in the Herbarium of Beijing Forestry University (BJFC). The total genome DNA was extracted using a modified protocol (Chen et al. Citation2014) and sent to Majorbio Company for next-generation sequencing using Illumina Hiseq Xten. About 2 Gb high quality, 2 × 150 base pairs PE reads were obtained from High-throughput sequencing. We used Map to Reference function in Geneious Prime 2019.0.4 to exclude nuclear and mitochondrial reads using published plastid genome of Acer truncatum (GenBank accession no. NC 037211.1) as the reference and used these filtered chloroplast reads for de novo assembly with Geneious Prime 2019.0.4 (Kearse et al. Citation2012). The assembled chloroplast sequences were then annotated using Plann (Huang and Cronk Citation2015) by referring to the relative group, and we perfected the annotation using Geneious.
The complete chloroplast genome of A. catalpifolium (Genbank accession no. XSJ012) was a circular molecule with a size of 156,225 bp in length, comprising a large single copy (LSC) region of 85,976 bp and a small single copy (SSC) region of 18,066 bp, which were separated by a pair of 26,092 bp inverted repeat (IR) regions. The sequence contained 135 unique genes, including eight rRNA, 40 tRNA, and 87 protein-coding genes. Among these genes, 15 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, trnA-UGC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron, and two genes (clpP and ycf3) had two introns. Most of the genes occurred in the single copy regions while seven protein coding genes (PCG; ndhB, rpl2, rpl23, rps7, rps12, ycf2, and ycf15), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23) were duplicated in the IR regions. The overall GC content of the chloroplast genome is 37.9% and those in the LSC, SSC, and IR regions are 36.1%, 32.2%, and 42.9% respectively.
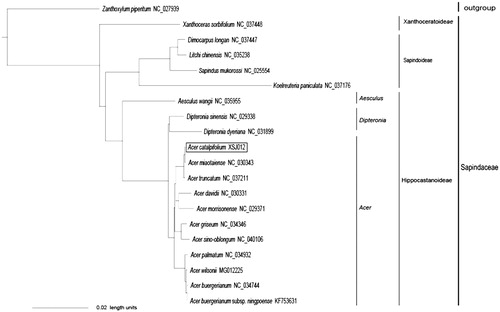
In order to understand its phylogenetic relationship within Sapindaceae, a phylogenetic analysis was performed based on reported complete chloroplast genome sequences of ten Acer species and eight other Sapindaceae species, and Zanthoxylum piperitum as the outgroup from NCBI. The Bayesian inference (BI) tree was reconstructed by MrBayes 3.2.3 (Ronquist and Huelsenbeck Citation2003). All the sequences were aligned using MAFFT v6.833 (Katoh et al. Citation2005), and the appropriate nucleotide substitution model for each sequence was according to the Akaike information criterion (AIC; Posada and Buckley Citation2004) using jModeltest (Posada Citation2008). In the phylogenetic tree (), the posterior probability (PP) of all branch nodes is 1.00. The phylogenetic analysis showed that A. catalpifolium is sister to A. miaotaiense, and the two species are then sister to A. truncatum. Meanwhile, 11 species of Acer and two species of Dipteronia are clustered into one big monophyletic clade, and the clade is then closely related to Aesculus wangii, which supported the present systematic placement of Aceraceae (Acer, Dipteronia) within subfamily Hippocastanoideae of Sapindaceae sensu lato.
Disclosure statement
The authors declare there is no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- APG (Angiosperm Phylogeny Group). 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Chen LY, Song MS, Zha HG, Li ZM. 2014. A modified protocol for plant genome DNA extraction. Plant Diversity Resour. 36:375–380.
- Fu LK. 1992. China plant red data book—rare and endangered plants. Beijing: Science Press.
- Harrington MG, Edwards KJ, Johnson SA, Chase MW, Gadek PA. 2005. Phylogenetic inference in Sapindaceae sensu lato using plastid matK and rbcL DNA sequences. Syst Bot. 30:366–382.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Pl Sci. 3:1500026.
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Ma Y, Chen G, Grumbine RE, Dao ZL, Sun WB, Guo HJ. 2013. Conserving plant species with extremely small populations (PSESP) in China. Biodivers Conserv. 22:803–809.
- Pan HL, Feng QH, Long TL, He F, Liu XL. 2014. Discussion on resource condition and protection technique for rare and endangered species in Sichuan Province. J Sichuan for Sci Tech. 35:41–46.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Wu BL, Long CL, Qing ST. 2018. Population structure and its quantity dynamics of Acer catalpifolium of Karst forest in Maolan National Natural Reserve. Acta Bot Bor-Occi Sin. 38:1918–1926.
- Zhang YY, Ma WB, Yu T, Ji HJ, Gao J, Li JQ, Gao S, Ke L. 2018. Population structure and community characteristics of Acer catalpifolium Rehd. Chin J Appl Environ Biol. 24:0697–0703.