Abstract
In this study, we described the complete mitochondrial genome of Alpine musk deer (Moschus chrysogaster) from China. It has a circular genome of 16,352 bp (GeneBank number: MK697349) with a higher A + T content of 62.06%, including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and 1 control region (D-loop), which was similar to typical vertebrate mitochondrial DNA. The phylogenetic analysis indicated that our sequence clustered with two Alpine musk deer subspecies from Sichuan, China. The results could provide more molecular information to support the conservation and genetic resources of Alpine musk deer.
The Alpine musk deer (Moschus chrysogaster) belongs to Moschidae, Artiodactyla, Mammalia, which is distributed in western China, including Tibet, Qinghai, Sichuan, and Gansu (Groves and Grubb Citation1989; Sheng Citation1992; Smith et al. Citation2009). Apline musk deer are generally solitary animals whose habitat is in coniferous forest, alpine shrubs at an altitude of 3000–5000 m (Meng et al. Citation2003). Due to the large amount of musk collection and habitat destruction, now, there are no more than 1000 wild individuals (Sheng Citation1998; Meng et al. Citation2003; Yang et al. Citation2003). It is now listed as a threatened species in the Red List by the International Union for the Conservation of Nature and Natural Resources (IUCN), and classified as national first class protected animal in China. The taxonomy of musk deer remains debatable (Flerov et al. Citation1952; Groves et al. Citation1995; Yang et al. Citation2013). However, Grove and Feng (Citation1986) believed that Tawny Musk Deer (Moschus fuscus) and Himalayan Musk Deer (Moschus chrysogaster) (Harris Citation2016) were subspecies of Apline Musk Deer.
Mitochondrial DNA is a powerful molecular tool for phylogenetic analysis and genetic diversity assessment of animals, there are currently only four complete mitochondrial genome (mitogenome) sequences reported about Alpine Musk Deer in GeneBank (Zhu et al. Citation2013). Here, we obtained a muscle sample of Apline Musk Deer from the forest public security of Baoxing county (N30°22′14.87″, E102°48′46.56″), which is located in the Hengduan Mountains between the Qinghai-Tibet Plateau and the Sichuan Basin. The sample was stored at zoology laboratory, Sichuan Agricultural University (Accession: sicau-lz-A596). Total genomic DNA was extracted by standard phenol–chloroform extraction (Sambrook and Russell Citation2001). We designed 11 primer pairs to amplify the complete Alpine Musk Deer mitochondrial genome.
The complete Alpine Musk Deer mitochondrial genome (GenBank number: MK697349) was 16,352 bp with an enriched A + T (62.06%). It is similar to most vertebrates, and the overall base composition was: A, 33.96%;T, 28.10%; G, 12.92%; and C, 25.02%. The complete sequence contained 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes (12S rRNA, and 16S rRNA), and one control region (D-loop). ND6 and eight tRNA genes were encoded on the light strand, other PCGs were distributed on the heavy strand. Most PCGs used the initiation codon ATG and ATT except ND3, which used ATA; 12 of the PCGs used complete (TAA) or incomplete (T– –) as stop codon; and the cytochrome b (Cytb) gene ended with AGA.
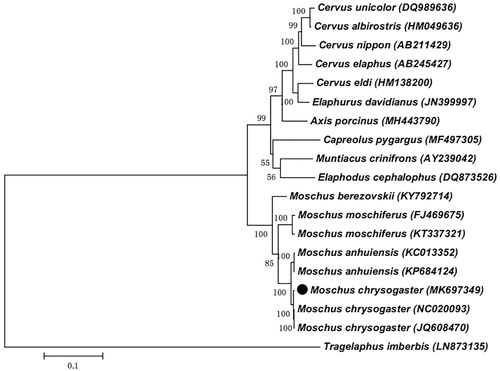
To further explore the taxonomic status of Alpine musk deer, a maximum likelihood (ML) phylogenetic tree was reconstructed in MEGA7.0, using an alignment comprising 19 complete mitogenome sequences from Artiodactyla, including two reported complete mitochondrial genome (mitogenome) sequences about Alpine Musk Deer and a Lesser Kudu (Tragelaphus imberbis) (LN873135) as an outgroup (). According to the ML tree, genus Moschidae and genus Cervidae clustered into two different branches. Our sequence was more closely related to the Anhui Musk Deer (Moschus anhuiensis) species. The result could provide more molecular information which can be used for the taxonomic classification and conservation.
Figure 1. Maximum likelihood (ML) phylogenetic tree of Alpine musk deer (Moschus chrysogaster) and the other 18 sequences from the order Artiodactyla with a sequence of Lesser Kudu (Tragelaphus imberbis) (LN873135) as an outgroup. Number at each node indicates the bootstrap support values. GenBank accession numbers are given in the brackets after the species name. The sequence characterized in this study is annotated with a black circle symbol.
Acknowledgements
We thank Qian Su, Xue Liu, Pu Zhao, Yuhan Wu and Mengmeng Dong for their assistance in the laboratory.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Flerov KK, Biron A, Cole ZS. 1952. Mammals: musk deer and deer. Moscow: USSR Academy of Sciences Press.
- Groves CP, Feng ZJ. 1986. The status of musk-deer from Anhui Province, China. Acta Theriol Sinica. 6:101–106.
- Groves CP, Grubb P. 1989. Relationships of living deer. In: Wemmer CM, editor. The biology and management of the Cervidae. Washington: Smithsonian Institution Press; p. 21–59.
- Groves CP, Wang YX, Grubb P. 1995. Taxonomy of musk-deer, genus Moschus (Moschidae, Mammalia). Acta Theriol Sin. 15:181–197.
- Harris R. 2016. Moschus chrysogaster. The IUCN Red List of Threatened Species. 2016:e.T13895A61977139. http://www.iucnredlist.org. Downloaded on 17 March 2019.
- Meng X, Yang Q, Feng Z, Xia L, Jiang Y, Wang P. 2003. Timing and synchrony of parturition in Alpine musk deer (Moschus sifanicus). Folia Zool. 52:39–50.
- Meng X, Yang Q, Xia L, Feng Z, Jiang Y, Wang P. 2003. The temporal estrous patterns of female alpine musk deer in captivity. Appl Anim Behav Sci. 82:75–85.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd edn. NewYork: Cold Spring Harbor Laboratory Press.
- Sheng HL. 1992. Chinese deer animal. Shanghai: East China Normal University Press.
- Sheng HL. 1998. Genus Moschus in China. Beijing: Science Press.
- Smith AT, Xie Y, Hoffmann RS, Lunde D, MacKinnon J, Wilson DE, Wozencraft WC. 2009. A guide to the mammals of China. Changsha: Hunan Education Publishing House Press.
- Yang Q, Meng X, Xia L, Feng Z. 2003. Conservation status and causes of decline of musk deer (Moschus spp.) in China. Biol Conserv. 109:333–342.
- Yang CZ, Xiang CZ, Zhang XY, Yue BS. 2013. The complete mitochondrial genome of the Alpine musk deer (Moschus chrysogaster). Mitochondrial DNA. 24:501–503.
- Zhu XX, Shi WB, Pan T, Wang H, Zhou LZ, Zhang B. 2013. Mitochondrial genome of the Anhui musk deer (Moschus anhuiensis). Mitochondrial DNA. 24:205–207.
