Abstract
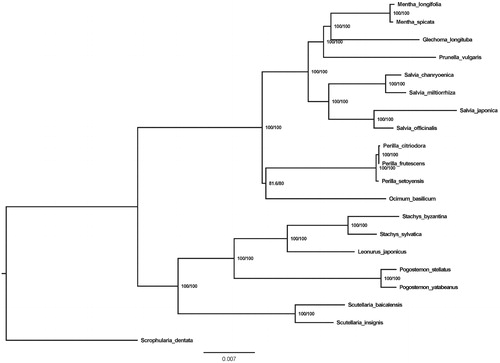
Glechoma longituba, a typical perennial clonal plant that is recorded in Chinese Pharmacopoeia 2015 version. In this study, the complete chloroplast genome (cpDNA) sequences of Glechoma longituba were reported. The cpDNA is 153,069 bp in length, contains a large single copy region (LSC) of 84,638 bp a small single copy region (SSC) of 17,520 bp, and two inverted repeat (IR) regions of 25,462 bp. The plastome contained 133 genes, including 87 protein-coding, 8 ribosomal RNA, and 37 transfer RNA genes. The overall GC content of the whole genome is 37.8% and the corresponding values of the LSC, SSC, and IR regions are 35.9, 32, and 43.1%, respectively. Phylogenetic analysis based on 19 chloroplast genomes indicates that a close relationship among Mentha longifolia, Mentha spicata, G. longituba, and Prunella vulgaris.
Clonal plants which ramets connected by rhizomes or stolons can share water, carbohydrates, and nutrients are widely distributed in all types of ecosystems, and dominate in many of them (de Kroon and Bobbink Citation1997; Herben and Hara Citation2003; Chen et al. Citation2010; Quan et al. Citation2018). Among them, Glechoma longituba, a typical perennial clonal plant that is recorded in Chinese Pharmacopoeia Commission Citation2010 version (State Pharmacopoeia Committee 2010). With the functions of eliminating damp and treating stranguria, clearing heat and removing toxins, dissipating blood stasis and diminishing swelling, and anti-microbial activity (Zhang et al. Citation2006). Therefore, it is significant to study this species for the ecosystem and pharmacology. Previous studies mainly focused on morphological and physiological aspects, the molecular genetic research of G. longituba is limited. Here, we assembled the complete chloroplast (cp) genome of G. longituba based on Illumina paired-end sequencing for further study.
The fresh leaves of a single individual of G. longituba were sampled from Xi’an Botanical Garden of Shaanxi Province (Shaanxi, China). Genomic DNA was extracted from the fresh leaves using the modified CTAB method (Doyle Citation1987). Total DNA was used for the shotgun library construction and the subsequent high-throughput sequencing on the Illumina HiSeq 2500 Sequencing System.
After quality-trimmed and assembled using MITObim v1.8 (Hahn et al. Citation2013) with the reference sequence of Scutellaria baicalensis (GenBank: MF521633.1). The genome was annotated using software Geneious v9.0.2 (Biomatters Ltd., Auckland, New Zealand) by aligning with the reference chloroplast genome. The circular plastid genome map was completed using the online program OGDRAW (Lohse et al. Citation2013). The annotated chloroplast genome sequence has been deposited into the GenBank with the accession number MK609928.
The total plastome length of G. longituba was 153,069 bp, with a large single copy (LSC; 84,638 bp), a small single copy (SSC;17,520 bp), and two inverted repeats (IRa and IRb; 25,462 bp each). The overall GC content was 37.8% (LSC: 35.9%; SSC: 32.0%; IRs: 43.1%) and the plastome contained 133 genes, including 87 protein-coding, 8 rRNA, and 37 tRNA genes. A total of 17 genes were duplicated in the inverted repeat regions including seven tRNA, four rRNA, and six protein-coding genes. The ycf1 gene was located at the SSC and IRa junction.
The neighbor-joining(NJ) phylogenetic tree was constructed with Model Finder (Kalyaanamoorthy et al. Citation2017) based on 19 complete chloroplast genome sequences of Lamiaceae and Scrophularia dentata (Scrophulariaceae) (NC_036942.1) as an outgroup (). The tree indicated that a close relationship among Mentha longifolia, Mentha spicata, G. longituba, and Prunella vulgaris. This complete chloroplast genome of G. longituba will provide information for future conservation genetic studies.
Figure 1. Neighbor-joining (NJ) phylogenetic tree based on 20 complete chloroplast genomes. Accession numbers: Mentha longifolia (NC_032054.1); Mentha spicata (NC_037247.1); Glechoma longituba (MF521633.1); Prunella vulgaris (NC_039654.1); Salvia chanryoenica (NC_040121.1); Salvia miltiorrhiza (NC_020431.1); Salvia japonica (NC_035233.1); Salvia officinalis (NC_038165.1); Perilla citriodora (NC_030755.1); Perilla frutescens (NC_030756.1); Perilla setoyensis (NC_030757.1); Ocimum basilicum (NC_035143.1); Stachys byzantina (NC_029825.1); Stachys sylvatica (NC_029824.1); Leonurus japonicus (NC_038062.1); Pogostemon stellatus (NC_031434.1); Pogostemon yatabeanus (NC_031433.1); Scutellaria baicalensis (NC_027262.1); Scutellaria insignis (NC_028533.1); Scrophularia dentata (NC_036942.1).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen JS, Lei NF, Dong M. 2010. Clonal integration improves the tolerance of Carex Praeclara to sand burial by compensatory response. Acta Oecologica. 36:23–28.
- Chinese Pharmacopoeia Commission, 2010. Pharmacopoeia of the People' s Republic of China, 2010 edition. Beijing: China Medical Science Press; p. 158–159.
- de Kroon H, Bobbink R. 1997. Clonal plant dominance under elevated nitrogen deposition, with special reference to Brachypodium pinnatum in chalk grassland. In: de Kroon H, van Groenendael J,, editors. The ecology and evolution of clonal plants. Leiden: Backhuys Publishers; p. 359–379
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-abaiting and iterative mapping approach. Nucleic Acids Res. 41:e129
- Herben T, Hara T. 2003. Spatial pattern formation in plant communities. In: Sekimura T, Noji S, Ueno N, Maini PK, editors. Morphogenesis and pattern formation in biological systems – experiments and models. Tokyo: Springer-Verlag; p. 223–235.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Model Finder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Quan JX, Zhang XY, Song SS, Dang H, Chai YF, Yue M, Liu X. 2018. Clonal plant Duchesnea indica Focke forms an effective survival strategy in different degrees of Pb-contaminated environments. Plant Ecol. 219:1315–1327.
- Zhang QJ, Yang XS, Zhu HY, Hao XJ. 2006. Chemical constituents and their pharmacological research progress in medicinal plants of Glechoma Linn.Chinese Trad Herb Drugs. 37:950–985.
