Abstract
Hydrotaea dentipes (Diptera: Muscidae) is one of the forensically important fly species and distributed worldwide. We describe the complete mitochondrial genome of the H. dentipes for the first time. It is a circular molecule of 16,070 bp in size, consisting of A (40.10%), G (9.20%), T (38.50%), and C (12.30%). The order and structure of the genes are similar to other Muscidae species. The mitochondrial genome (mitogenome) is composed of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), two ribosomal RNA genes (rRNA), and a non-coding AT-rich region. Most PCGs used the canonical putative codon to start and stop.
Hydrotaea dentipes (Fabricius, 1805), is distributed widely in Asia and Canada (Chen Citation2013). They commonly colonize garbage, feces, and carcasses; the larvae usually are found in faeces and their growth into pupa only need a few days in the summer. Thus, bionomics of H. dentipes making it was detrimental in animal husbandry and public health (Mehlhorn Citation2008). Mitogenome has been widely used for species identification as a molecular marker (Shang et al. Citation2019), estimations of evolutionary relationships in phylogenetic analyses, and population genetics.
Specimens of H. dentipes were collected in August 2018 in Lhasa city (29°36’N, 91°06’E), China. Total genomic DNA was extracted from the thorax muscle and is currently stored at Guo's Lab (Changsha city, 28°12’N, 112°58’E). The H. dentipes mitogenome has been submitted to GenBank with the accession number of MK820719.
The complete mitogenome of H. dentipes has been obtained from high-throughput sequencing on complete mitochondrial DNA. The circular mitogenome of H. dentipes is 16,070 bp in length, containing the classical Diptera’s structure of 37 genes, and a non-coding AT-rich region. To make sure the authenticity of gene boundaries, all PCGs were compared with other muscid mitochondrial sequences underperformed in MEGA 7.0 (Kumar et al. Citation2016). Twenty-one tRNAs of H. dentipes can be folded into the typical clover-leaf structure, whereas the tRNA-Ser (trnS1) has a special clover-leaf structure without a dihydrouridine (DHU) arm and is replaced by a simple loop.
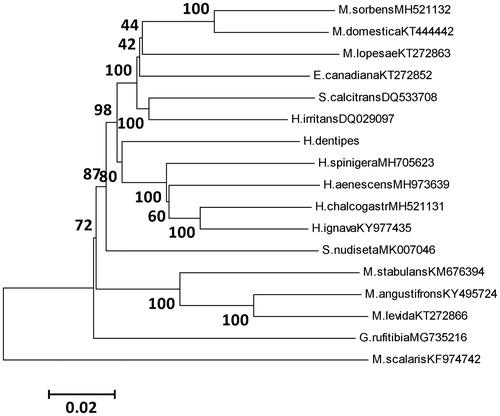
We conducted a phylogenetic analysis using the 13 PCGs from 16 species of Muscidae species, along with Megaselia scalaris from family Phoridae as outgroup species, and the tree was built by using neighbour-joining method (). The tree shows H. dentipes belongs to Azeliinae, and the Azeliinae is closely related to Muscinae, which was consistent with the study by Ren et al. (Citation2019).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen LS. 2013. Necrophagous flies in China. Guiyang (China): Guizhou Science and Technology Press.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Mehlhorn H. 2008. Encyclopedia of parasitology – Hydrotaea. New York: Springer-Verlag Berlin Heidelberg.
- Ren LP, Shang YJ, Yang L, Shen X, Chen W, Wang Y, Cai JF, Guo YD. 2019. Comparative analysis of mitochondrial genomes among four species of muscid flies (Diptera: Muscidae) and its phylogenetic implications. Int J Biol Macromol. 127:357–364.
- Shang Y, Ren L, Chen W, Zha L, Cai J, Dong J, Guo Y. 2019. Comparative mitogenomic analysis of forensically important sarcophagid flies (Diptera: Sarcophagidae) and implications of species identification. J Med Entomol. 56:392–407.