Abstract
The mitochondrial genome of Odorrana graminea was sequenced and analyzed. The complete mitochondrial genome is 18,106 bp in length, including 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 21 transfer RNA genes, and a control region (D-loop). All PCGs are initiated by ATN codons, except for GTG for COI and ND5. Five PCGs use a common stop codon of TAA or TAG, whereas COI terminats with AGG as stop codon; ND6 with AGA; the other six ends with an incomplete stop codon (a single stop nucleotide T). The analysis results based on Bayesian inference method provide a useful resource for the phylogenetic studies of superfamily Ranoidea.
The large odorous frog O. graminea is an arboreal, nocturnal frog living near noisy streams and waterfalls in mountainous areas of southern China (Shen Citation2018). However, O. graminea is threatened by human activities, such as habitat destructions, excessive captures, and pollutions (Fei et al. Citation2012). Therefore, it has been listed in The IUCN Red List (https://www.iucnredlist.org/species/58608/11808913). In this study, we determined the complete mitochondrial DNA (mtDNA) sequence of O. graminea (GenBank accession no. MN417331) for the protection of this species.
The specimen of O. graminea was collected from Guanshan National Nature Reserve in Yifeng County, Jiangxi Province, China (N28°30′–28°40′, E114°29′∼114°45′), and was deposited in Guangdong Institute of Applied Biological Resources (No. 2018098). Total genomic DNA was extracted with the TaKaRa MiniBEST Universal Genomic DNA Extraction Kit (Takara, Beijing, China) following the kit’s instructions. The high-quality genomic DNA were generated on an Illumina HiSeq4000 using sequencing protocols provided by the manufacturer (Illumina, Inc., San Diego, CA, USA).
The complete mitochondrial genome of O. graminea is 18,106 bp in length with 29.40% A, 27.96% C, 14.75% G, and 27.89% T. The genome is comprised of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 1 control region, similar to typical vertebrate mtDNA. The total length of 22 tRNA genes is 1523 bp, ranging from 64 bp (tRNACys) to 73 bp (tRNALeu (UUR) and tRNAAsn). The total length of 13 protein-coding genes is 11,295 bp. The start codons are ATG for ND1, COII, ATP8, COIII, ND3, ND4L, ND4, ND6, and CYTB while ATT for ND2, GTG for COI and ND5 and ATA for ATP6. COI terminats with AGG as stop codon; ND6 with AGA; ATP8, ND4L, ND5 and CYTB with TAA; ND4 with TAG, and other six PCGs end with an incomplete stop codon (a single stop nucleotide T). The two ribosomal RNA genes including 12SrRNA (933 bp) and 16SrRNA (1585 bp) are commonly located between tRNAPhe and tRNALeu (UUR). The control region is 2568 bp in length with 63.86% A + T content. Within the complete mitochondrial genome of O. graminea, there are five reading frame overlaps (The tRNAGln and tRNAMet share one nucleotide; OL and tRNACys share three nucleotides; COI and tRNASer share nine nucleotides; ATP8 and ATP6 share one nucleotide; ND4L and ND4 share seven nucleotides).
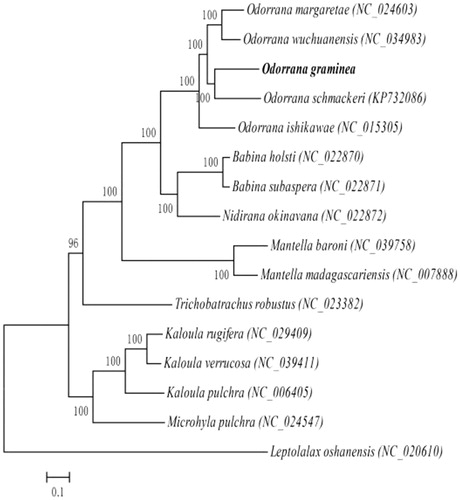
The phylogenetic analysis of the superfamily Ranoidea was conducted with other 14 Ranoidea mitogenomes and Leptolalax oshanensis (Xiang et al. Citation2013) mt genome as the outgroup with Bayesian inference (BI). The best-fitted substitution model was determined in MUSCLE v.3.8.31 (Edgar Citation2004) by Akaike Information Criterion (AIC). The BI was conducted in MrBayes 3.2.6 (Ronquist et al. Citation2012). As shown in , the phylogenetic positions of these 16 mt genomes are successfully resolved with high bootstrap supports. The result indicates O. graminea and O. schmackeri (Bu et al. Citation2016) are closest and the genetic distance between Ranidae and Mantellidae are closer than others.
Figure 1. Phylogenetic relationships (Bayesian inference) among the species of the superfamily Ranoidea based on the mitochondrial genome nucleotide sequence of the 13 protein-coding genes. Numbers beside the nodes are percentages of 1000 bootstrap values. Leptolalax oshanensis was used as outgroup.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bu XJ, Zhang LJ, He KX, Jiang YM, Nie LW. 2016. The complete mitochondrial genome of the Odorrana schmackeri (Anura, Ranidae). Mitochondr DNA B. 1(1):162–163.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Fei L, Ye CY, Jiang JP. 2012. Colored atlas of Chinese amphibians and their distributions. Chengdu: Sichuan Publishing House of Science and Technology.
- Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shen JX. 2018. Ultrasonic vocalization in amphibians and the structure of their vocal apparatus. Handb Behav Neurosci. 25:481–491.
- Xiang TM, Wang B, Liang XX, Jiang JP, Li C, Xie F. 2013. Complete mitochondrial genome of Paramegophrys oshanensis (Amphibia, Anura, Megophryidae). Mitochondr DNA. 24(5):472–474.
