Abstract
Gelsemium elegans, endemic to southern Asia, is a highly toxic plant with various medicinal functions. The complete mitochondrial genome of G. elegans was sequenced and assembled in this study. The genome is 405,990 bp in length and contains 37 protein-coding genes, 20 tRNA genes, and 3 rRNA genes. The phylogenetic tree based on 15 protein-coding genes common to six mitochondrial genomes in Gentianales support that G. elegans of Gelsemiaceae is sister to Apocynaceae.
Gelsemium elegans (Gardner & Champ.) Benth., also known as heartbreak grass or ‘Duan Chang Cao’ in Chinese, is a highly poisonous vine in Gelsemiaceae. It is indigenous to south and southeast Asia. Gelsemium extract contains toxic alkaloids such as gelsemine, gelsenicine, gelsevirine, and koumine (Zhang et al. Citation2015). Intoxication of G. elegans could cause paralysis, asphyxia, convulsions, and even death due to multiple organ failure in severe conditions (Zhou et al. Citation2017). Previous studies demonstrated that G. elegans had anti-inflammatory, immunomodulating, analgesic, anxiolytic, anti-tumor, and neuropathic pain-relieving properties (Liu et al. Citation2019). In this study, we report the first complete mitochondrial genome of G. elegans which will provide essential genetic data for further genetic improvement and effective use of this medicinally important species.
The leaves of G. elegans were collected from Liuzhou, Guangxi, China (GPS: E109°14'43'', N24°20'40'', Voucher No. ZSTU03201, deposited at Herbarium of Zhejiang University). DNA was extracted from its silica dried leaves using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA). The total genomic sequences were generated using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA). In total, about 9.2 G high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. Firstly, the chloroplast genome of G. elegans was firstly assembled following Liu et al. (Citation2017, Citation2018). Then, sequence of G. elegans plastome and a sequence of ATPase subunit 1 (Accession: KF541337) were set as reference in NOVOPlasty (Dierckxsens et al. Citation2017) for mitochondrion genome de novo assembly. Finally, aligning and annotation were conducted by BLAST, GeSeq (Tillich et al. Citation2017) and GENEIOUS Prime (Biomatters Ltd., Auckland, New Zealand).
The complete mitochondrial genome of G. elegans (Accession: MN388837) has a length of 405,990 bp with a typical circle structure and a GC content of 44.4%. The genome contains 37 protein-coding genes, including ones for NADH dehydrogenase (nad1, 2, 3, 4, 4L, 5, 6, 7, 9), cytochrome c oxidase (cox1, 2, 3), cytochrome c biogenesis (ccmB, C, Fn, Fc), apocytochrome b (cob), ATP synthase (atp1, 4, 8, 9), ribosomal proteins (rpl2, 5, 10 and rps1, 3, 4, 7, 10, 12, 13, 19), succinate dehydrogenase(sdh3, 4), maturase and membrane transporter (mttB and matR). Additionally, the genome also contains 20 tRNA genes coding for 17 amino acids, 3 rRNA genes (rrn5, 18, 26).
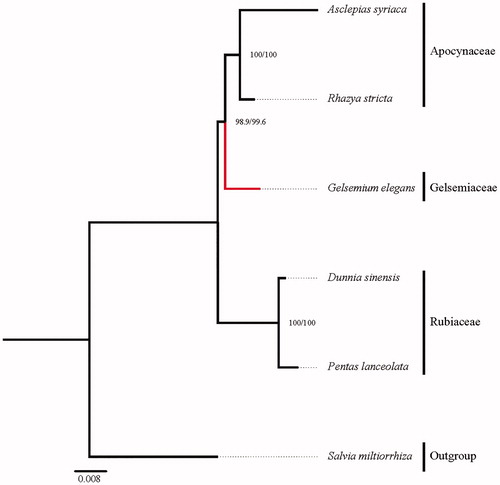
Fifteen mitochondrial specific protein-coding genes (atp1, atp4, atp8, atp9, ccmB, ccmC, cob, cox2, mttB, nad3, nad4, nad7, rpl5, rps13, rps3) from six species were selected to study the phylogenetic placement of G. elegans in Gentianales. The sequence alignment was conducted by MAFFT v7.3 (Katoh and Standley Citation2013). The software IQtree (Nguyen et al. Citation2015) was used to draw the phylogenetic tree with 5000 bootstrap replicates and TVM + F + G4 model. The phylogenetic tree exhibited that G. elegans of Gelsemiaceae was closely related to Apocynaceae in Gentianales ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Figure 1. The best maximum-likelihood (ML) phylogram inferred from 6 plant mt genomes based on 15 protein-coding genes. Accession numbers are as follows: Asclepias syriaca (KF541337), Rhazya stricta (KJ485850), Salvia miltiorrhiza (KF177345), Pentas lanceolate (KY492150, KY637224, KY637398, KY637630, KY637974, KY638032, KY638090, KY638148, KY638264, KY638495, KY638957, KY639015, KY639073, KY639131, KY639229), and Dunnia sinensis (KY492163, KY637237, KY637411, KY637643, KY637987, KY638045, KY638103, KY638161, KY638277, KY638506, KY638970, KY639028, KY639086, KY639144, KY639242).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in Performance and Usability. Mol Biol Evol. 30:772–780.
- Liu L, Li P, Zhang HW, Worth J. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T. Sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Forest Res-JPN. 23:318–323.
- Liu L, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu C. 2017. The complete chloroplast genome of Chinese Bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Liu Y, Tang Q, Cheng P, Zhu M, Zhang H, Liu J, Zuo M, Huang C, Wu C, Sun Z, et al. 2019. Whole-genome sequencing and analysis of the Chinese herbal plant Gelsemium elegans. Acta Pharm Sin B. accessed on August 30th, 2019
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucl Acids Res. 45:W6–W11.
- Zhang QP, Wang ZT, Chou GX. 2015. Preparative separation of four alkaloids from Gelsemium elegans by high-speed counter-current chromatography. Chin Herb Med. 7:267–272.
- Zhou Z, Wu L, Zhong Y, Fang X, Liu Y, Chen H, Zhang W. 2017. Gelsemium elegans poisoning: a case with 8 months of follow-up and review of the literature. Front Neurol. 8:204.
