Abstract
We report a complete chloroplast (cp) genome of a Cymbidium tortisepalum hermaphrodite. The cp genome is 149,764 bp in length, with a large single-copy region (LSC) of 85,257 bp and a small single-copy region (SSC) of 13,317 bp, which were separated by a pair of 25,595 bp inverted repeat regions (IRs). The genome contained 130 genes, with 111 unique genes, including 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The overall GC content is 37.11% with the values of the LSC, SSC, and IR regions are 34.44%, 29.67%, and 43.49%, respectively. Further, phylogenetic analysis suggested that the plastome of C. tortisepalum hermaphrodite is close to published C. sinense (NC021430.1), C. kanran (KU179435.1), C. tortisepalum (NC021431.1), and C. ensifolium (NC028525.1) plastomes.
Cymbidium tortisepalum Lindl. with common name ‘Lianban Lan’, is an evergreen species native to China (Chen et al. Citation2009). The species contains many varieties with mutant floral and plant phenotypes, which have high ornamental value. Cymbidium tortisepalum hermaphrodite, with intact female and male organs is one of these famous phenotypes. This type blooms in Spring and is widely cultivated in southeast Asian countries. Here, we reported the complete chloroplast genome of C. tortisepalum hermaphrodite and figure out its phylogenetic position in genus Cymbidium.
The complete plastome sequence of C. tortisepalum hermaphrodite was sequenced and assembled from Illumina reads. The specimen was collected from Dali, Yunnan, China (N 25°25′, E100°00′) and stored in Fujian Agriculture and Forestry University (specimen voucher X.K. Ma 003). After DNA extraction from fresh leaves, the library was constructed and DNA was sequenced using the high-throughput sequencing (PE 150) on an Illumina Hiseq 2500 platform (Illumina, San Diego, CA). Totally 8 Gb raw reads were generated. The filtered reads were assembled using GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle) with a list of complete chloroplast genomes in orchid family as the reference. The assembled chloroplast genome was viewed and edited using Bandage software (Wick et al. Citation2015), then was annotated using Geneious R11.15 (Kearse et al. Citation2013; Markowitz et al. 2012). The structural features of annotations were visualized using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/).
The new assembled complete chloroplast genome (GenBank accession NO. MN497245) of C. tortisepalum hermaphrodite is 149,764 base pairs (bp) in length, containing a large single-copy (LSC) region of 85,257 bp, and a small single-copy (SSC) region of 13,317 bp, which were separated by two inverted repeat (IR) regions of 25,595 bp. The plastome possesses total 130 genes, with 111 unique genes (including 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes). Among all of these genes, seven protein-coding genes (i.e. ndhB, rpl2, rps12, rps19, rps7, rpl23, and ycf2), four rRNA genes (i.e. 4.5S, 5S, 16S, and 23S rRNA), and eight tRNA genes (i.e. trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) occur in double copies. The GC-content of the whole plastome is 37.11%, with the corresponding values of the LSC, SSC, and IR regions are respectively 34.44%, 29.67%, and 43.49%.
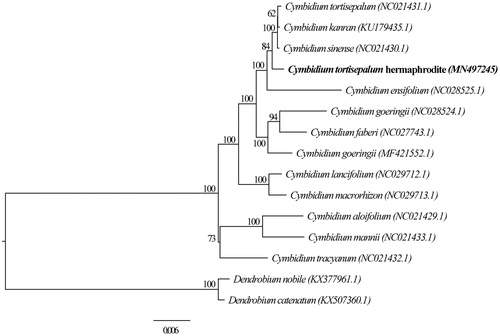
To further investigate its phylogenetic position, a ML tree was constructed based on plastome sequences of 12 Cymbidium species and two outgroup Dendrobium species using IQ-tree (Nguyen et al. Citation2015) with 1000 bootstrap replicates. We aligned all 15 sequences using Muscle (Edgar Citation2004). BMGE tools (Criscuolo and Gribaldo Citation2010) were then used to prune the aligned sequences. The ModelFinder module in IQ-tree was used to search the optimal nucleotide substitution model. Finally, the best model TVM + F+R2 was chosen to construct the phylogenetic tree according to BIC criteria. Our phylogenetic analysis suggested that the plastome of C. tortisepalum hermaphrodite is closely related to published plastomes of Cymbidium sinense, Cymbidium kanran, C. tortisepalum, and Cymbidium ensifolium with 100% bootstrap support (). This newly reported plastome of C. tortisepalum hermaphrodite provides a good resource for studying Cymbidium phylogenetic relationships and their plastome evolution.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen X, Liu Z, Zhu GH, Lang KY, Ji ZH, Luo YB, Jin XH, Cribb PJ. 2009. Orchidaceae, flora of China. St. Louis (MO): Science Press & Missouri Botanical Garden Press.
- Criscuolo A, Gribaldo S. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10(1):210.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Lohse M, Drechsel O, et al. 2013. Organellar genome DRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Markowitz S, Duran C, Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.