Abstract
The rhizome of Atractylodes macrocephala is one of the most commonly used herbs in China. In this paper, we presented the complete chloroplast genome of A. macrocephala. The chloroplast genome of A. macrocephala is 153,256 bp in length as the circular, which harbors a large single-copy (LSC) region 84,291 bp, a small single-copy (SSC) region of 18,675 bp and separated by a pair of inverted-repeat (IR) regions of 25,145 bp for each one. The overall nucleotide content of the chloroplast genome is 37.7% GC content. This chloroplast genome contains 125 genes, which includes 88 protein-coding genes (PCGs), 29 transfer RNA (tRNAs) and 8 ribosome RNA (rRNAs). Phylogenetic implications based on chloroplast genomes of 16 the family Compositae plant species indicated that Atractylodes macrocephala was closely related to Atractylodes lancea in the family Compositae by the Maximum-Likelihood (ML) method.
The rhizome of Atractylodes macrocephala (Bai-Zhu in Chinese) has been used in Traditional Chinese Medicine (TCM) for about 2,000 years in China (Shan Citation1981). It was awarded the title as the first herb of invigorating qi and strengthening spleen, which is also as one of eight well-known medicinal herb specialties in Zhejiang province and the most famous and much more expensive than other regions (Hu et al. Citation2019). The rhizome of A. macrocephala possesses various pharmacological effects that involves anti-mutation, anti-tumor, anti-aging, promotion of cellular growth and so on (Li et al. Citation2012). Now, the study of A. macrocephala mainly focuses on the composition and activity, little molecular biology and bioinformatics information about this species. In this paper, the chloroplast genome of A. macrocephala was presented that can provide some genome information and data for the family Compositae plant, also can be TCM business to the world in further.
The fresh root of Atractylodes macrocephala as the sample was collected from herb market near Zhejiang Chinese Medical University that located at Hangzhou, Zhejiang, China, 30.09 N, 119.89E. The chloroplast genomic DNA of A. macrocephala was extracted from the fresh root using the modified CTAB method and stored in Zhejiang Chinese Medical University (No. SCMC-ZJU-TCM-03). The NEB Next UltraTM II DNA Library Prep Kit (NEB, BJ, and CN) was used to purify the chloroplast DNA, which used by using sequencing. The low-quality reads and adapters was removed using FastQC software (Andrews Citation2015) for quality control. The chloroplast of A. macrocephala genome was assembled and annotated using the MitoZ software (Meng et al. Citation2019). The physical map of the chloroplast A. macrocephala genome used OGDRAW software (Greiner et al. Citation2019) to draw. The information and sequence of chloroplast genome had been deposited in GenBank with the accession number MN0446711.
The chloroplast genome of Atractylodes macrocephala is 153,256 bp in length which is the circular with a characteristic quadripartite structure. It harbors a large single-copy (LSC) region of 84,291 bp, a small single-copy (SSC) region of 18,675 bp and separated by a pair of inverted repeat (IR) regions of 25,145 bp each. The chloroplast genome of A. macrocephala contains 125 genes, which includes 88 protein-coding genes (PCGs), 29 transfer RNA genes (tRNAs) and 8 ribosomal RNA genes (rRNAs). In each IR region, the total of 17 genes, which included 8 PCG genes species (rpl12, rpl23, ycf2, ndhB, rps, rps12, ycf15 and ycf1), 5 tRNA genes species (trnI-CAU, trnL-CAA, trnV-GAC, trnR-ACG and trnN-GUU) and 4 rRNA genes species (rrn16, rrn23, rrn4.5 and rrn5) were found duplicated. The overall nucleotide content of the chloroplast genome: 31.0% A (Adenine), 31.3% T (Thymine), 18.6% C (Cytosine), 19.1% G (Guanine), and 37.7% GC content.
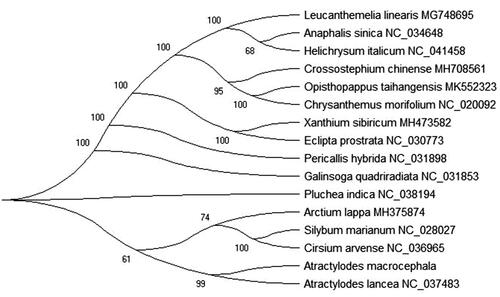
To further investigate Atractylodes macrocephala phylogenetic implications, the Maximum-Likelihood (ML) was used to construct phylogenetic tree and based on chloroplast genomes of 16 the family Compositae plant species. ML phylogenetic tree analysis used the MEGA X software (Kumar et al. Citation2018) and performed using 2,000 bootstrap values replicate at each node. All of the nodes were inferred with strong support by the ML methods. The final NJ phylogenetic tree was edited using the iTOL version 4.0 (https://itol.embl.de/) (Letunic and Bork Citation2016). Phylogenetic implications indicated that Atractylodes macrocephala was closely related to Atractylodes lancea in phylogenetic relationship (). This paper can provide much information and data for the family Compositae plant and more can be TCM business to the world.
Disclosure statement
The authors have declared that no competing interests exist.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Hu L, Chen X, Yang J, Guo L. 2019. Geographic authentication of the traditional chinese medicine Atractylodes macrocephala koidz. (Baizhu) using stable isotope and multielement analyses. Rapid Commun Mass Sp. 33(22):1703–1710.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evolution. 35(6):1547–1549.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v4: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Li X, Lin J, Han W, Mai W, Wang L, Li Q, Lin M, Bai M, Zhang L, Chen D. 2012. Antioxidant ability and mechanism of rhizoma Atractylodes macrocephala. Molecules. 17(11):13457–13472.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Shan ZJ. Check and punctuate on Shennong’s Classic of Herbology. Hefei (China): Wannan Medial College Press; 1981. p. 48.