Abstract
The complete chloroplast genome from Euonymus szechuanensis, a rare deciduous shrub of the Celastraceae endemic to China, which is classified as vulnerable on the list of Threatened species in China because species diversity decreased sharply, is determined in this study. The whole chloroplast genome sequence of Euonymus szechuanensis has been characterized by Illumina pair-end sequencing. The circular genome is 157,465 bp long, containing a large single-copy region (LSC) of 86,401 bp and a small single-copy region (SSC) of 18,472 bp, which are separated by a pair of 26,296 bp inverted repeat regions (IRs). It encodes a total of 136 genes, including 84 protein-coding genes (76 PCG species), 44 tRNA genes (30 tRNA species), and 8 ribosomal RNA genes (4 rRNA species). Most of the gene species occur as a single copy, while 19 gene species occur in double copies. The overall A + T content of the circular genome is 62.8%, while the corresponding values of the LSC, SSC and IR regions are 65.0, 68.4 and 57.3%, respectively. Phylogenetic analysis suggested that E. szechuanensis was relatively close to Euonymus schensianus among the species analyzed. This complete chloroplast genome will provide valuable insight into conservation and restoration efforts for this rare and Threatened species.
Euonymus szechuanensis is a species of the Celastraceae, a deciduous shrub mainly distributed in western China. This plant not only has ornamental value as garden plants but also can be used as a traditional Chinese medicine for the disintoxicating effect (Zhu et al. Citation2002). However, it has been classified as “rare and Endangered” in the List of Threatened Species in China, mainly because of its sharp decrease in species diversity. Therefore, this species needs to be better studied and prioritized as a conservation target. In the present study, we assembled the complete chloroplast genome sequence of Euonymus szechuanensis, which is an important resource for future studies on molecular markers to monitor population dynamics of the protected species.
Genomic DNA was extracted from fresh leaves of an individual of Euonymus szechuanensis collected from Ziyang County (Sichuan Province, China). The specimen was deposited at Xianyang Normal University (Accession number: LHZ-2018-QLM17-21). A genomic shotgun library with an insert size of 600 bp was prepared and then this library was sequenced and 150 bp paired-end reads were generated. Ten million high-quality reads were mapped to most of the published Celastraceae chloroplast genomes as references using BWA (Li and Durbin Citation2009). Secondly, we assembled these reads into complete chloroplast genomes using Velvet (Zerbino and Birney Citation2008). The assembled chloroplast genome was annotated using the online annotation tool DOGMA (Wyman et al. Citation2004) and further corrected the annotation with Geneious (Kearse et al. Citation2012). The complete chloroplast genome sequence with gene annotations were submitted to GenBank under the accession numbers of MH853828. The chloroplast sequence of the Euonymus szechuanensis was 157,465 bp. It possessed a typical quadripartite structure with two identical copies of a large inverted repeat separated by large and small single-copy regions. The large single-copy region (LSC) in E. szechuanensis was 86,401 bp and the small single-copy region (SSC) was 18,472 bp, which were separated by a pair of 26,296 bp inverted repeat regions (IRs). The circular genome contained 136 genes, including 84 protein-coding genes (76 PCG species), 8 ribosomal RNA genes (4 rRNA species), and 44 tRNA genes (30 tRNA species). Most of the gene species occurred in a single copy, while 19 gene species occurred in double copies, including 4 rRNA species (4.5S, 5S, 16S, and 23S rRNA), 7 tRNA species and 5 PCG species. The overall A + T content of the circular genome was 62.8%, while the corresponding values of the LSC, SSC, and IR regions were 65.0, 68.4, and 57.3%, respectively.
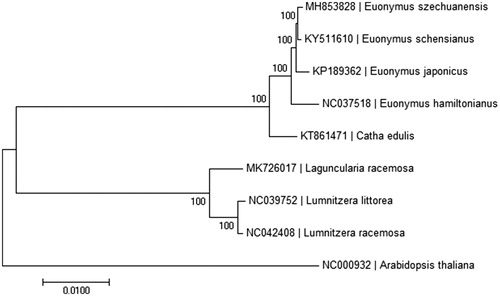
In order to ascertain its phylogenetic placement within the family Celastraceae, a neighbor-joining phylogenetic tree was constructed based on chloroplast genomes for a panel of 9 species with the program MEGA6 (Tamura et al. Citation2013). The result indicated that Euonymus szechuanensis was found to be relatively closely related to Euonymus schensianus compared to other species of Euonymus genera in Celastraceae (). We believe that it will provide valuable insight into conservation and evolutionary histories for this rare but important species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinforma. 25(14):1754–1760.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhu JB, Wang MG, Wu WJ, Zq J, Hu ZN. 2002. Insecticidal sesquiterpene pyridine alkaloids from Euonymus species. Phytochemistry. 61(6):699–704.