Abstract
We presented two complete chloroplast genomes of cold resistance coffee trees (Coffea arabica L.), named as CH1 and CH2 cultivated in Jeju island, Korea. Their length is 155,187 bp long (GC ratio is 37.4%) and has four subregions: 85,160 bp of large single copy (LSC; 35.4%) and 18,135 bp of small single copy (SSC; 31.3%) regions are separated by 25,946 bp of inverted repeat (IR; 43.0%) regions including 131 genes (86 protein-coding genes, eight rRNAs, and 37 tRNAs). Like sequenced coffee chloroplast genomes, number of sequence variations of CH1 and CH2 against TY1 chloroplast genomes are zero and two, respectively.
Coffea arabica L. is one of the major economical plants occupying 70% of world coffee production (Lashermes et al. Citation1999; O’brien and Kinnaird Citation2003). Seven coffee chloroplast genomes are available presenting low level of intraspecies variations (Samson et al. Citation2007; Min, Kim et al. Citation2019; Park, Kim, Xi, Heo Citation2019a, Citation2019b; Park, Kim, Xi, Nho et al. Citation2019; Park, Xi, Kim, Heo, et al. Citation2019). Cold resistance feature of coffee tree can expand cultivation area beyond ‘Bean Belt’ (Bentley and Baker Citation2000). Here, chloroplast genomes of two cold resistance coffee trees (CH1 and CH2) which can survive until –2 °C in Jeju island, Korea, without greenhouse were sequenced.
Total DNA of coffee trees (CH1 and CH2; J. Park, IBS-00013 and IBS-00014, respectively, InfoBoss Cyber Herbarium (IN); 33.239325 N, 126.287332E) was extracted from fresh leaves by using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Sequencing was performed using HiSeq2000 at Macrogen Inc., Korea. Filtered raw reads by Trimmomatic 0.33 (Bolger et al. Citation2014) were subjected to de novo assembly and sequence confirmation processes conducted with Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation with C. arabica TY1 chloroplast (MK862266; Min, Kim et al. 2019).
Both chloroplast genomes of CH1 and CH2 (GenBank accessions are MN894550 and MN894551, respectively) are identical: its length is 155,189 bp long (GC ratio is 37.4%) and has four subregions: 85,160 bp of large single copy (35.4%) and 18,135 bp of small single copy (31.3%) regions are separated by 25,946 bp of inverted repeat (IR; 43.0%). It contains 131 genes (86 protein-coding genes, eight rRNAs, and 37 tRNAs); 19 genes (8 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions.
CH1 and CH2 chloroplast genomes have six insertions and deletions (INDELs) against CH3, another cold hardness coffee (Park, Kim Xi, Kim Citation2019b) and two INDELs against TY1 chloroplast. It is similar to those of Camellia japonica (Park, Kim, Xi, Oh, et al. Citation2019b), Cucumis melo (Zhu et al. Citation2016), Chenopodium quinoa (Maughan et al. Citation2019), Abeliophyllum distichum (Park, Kim, Xi, Jang et al. Citation2019), Salix koriyanagi (Park, Kim, Xi, Kwon et al. Citation2019), Populus alba x Populus glandulosa (Park, Kim, Xi, Kwon et al. Citation2019), and Marchantia polymorpha (4 SNPs; Kwon et al. Citation2019) and smaller than those of other chloroplasts of Pyrus ussuriensis (Cho et al. Citation2019), Pseudostellaria palibiniana (Kim, Heo et al. Citation2019), Aconitum coreanum (Kim, Yi, et al. Citation2019), Artemisia fukudo (Min, Park et al. Citation2019), Goodyera schlechtendaliana (Oh et al. Citation2019), Dysphania pumilio (Park and Kim Citation2019), Nymphaea alba (Park, Kim, Kwon, Nam, et al. Citation2019), Liriodendron tulifipera (Park, Kim, Kwon, Xi, et al. Citation2019), Duchesnea chrysantha (Park, Kim, Lee Citation2019), Illicium anisatum (Park, Kim, Xi Citation2019a), Salix koriyanagi (Park, Kim, Xi Citation2019b), Viburnum erosum (Choi et al., Citation2020), Abeliophyllum distichum ( Min et al., Citation2019; Park et al., 2019), and Camellia japonica (Park, Kim, Xi, Oh et al. Citation2019a).
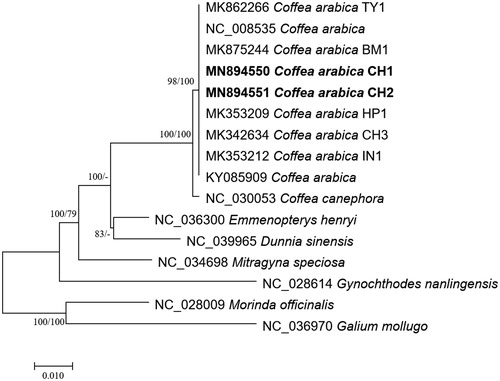
Ten Coffea and four chloroplast genomes of Rubiaceae as outgroup species were used for constructing bootstrapped neighbor joining and maximum likelihood trees using MEGA X (Kumar et al. Citation2018) after multiple sequence alignment by MAFFT 7.450 (Katoh and Standley Citation2013). Phylogenetic trees show that CH1 and CH2 are clustered in the C. arabica clade ().
Figure 1 Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of six Coffea and four Rubiaceae complete chloroplast genomes: nine Coffea arabica (MN894550 and MN894551 in this study, MK875244, MK862266, MK353212, NC_008535, KY085909, MK342634, and MK353209), Coffea canephora (NC_030053), Mitragyna speciosa (NC_034698), Dunnia sinensis (NC_039965), Emmenopterys henryi (NC_036300), and Gynochthodes nanlingensis (NC_028614). Phylogenetic tree was drawn based on maximum likelihood tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Bentley JW, Baker PS. 2000. The Colombian Coffee Growers’ Federation: organised, successful smallholder farmers for 70 years. ODI.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Cho MS, Kim Y, Kim SC, Park J. 2019. The complete chloroplast genome of Korean Pyrus ussuriensis Maxim. (Rosaceae): providing genetic background of two types of P. ussuriensis. Mitochondrial DNA Part B. 4(2):2424–2425.
- Choi Y G, Yun N, Park J, Xi H, Min J, Kim Y, Oh S-H. 2020. The second complete chloroplast genome sequence of the Viburnum erosum (Adoxaceae) showed a low level of intra-species variations. Mitochondrial DNA Part B. 5(1):271–272. doi:10.1080/23802359.2019.1698360.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim Y, Heo KI, Park J. 2019. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): intraspecies variations based on geographical distribution. Mitochondrial DNA Part B. 4(1):1310–1311.
- Kim Y, Yi JS, Min J, Xi H, Kim DY, Son J, Park J, Jeon JI. 2019. The complete chloroplast genome of Aconitum coreanum (H. Lév.) Rapaics (Ranunculaceae). Mitochondrial DNA Part B. 4(2):3404–3406.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution. 35(6):1547–1549.
- Kwon W, Kim Y, Park J. 2019. The complete chloroplast genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. and Boisselier: low genetic diversity between Korea and Japan. Mitochondrial DNA Part B. 4(1):959–960.
- Lashermes P, Combes M.-C, Robert J, Trouslot P, D’Hont A, Anthony F, Charrier A. 1999. Molecular characterisation and origin of the Coffea arabica L. genome. Mol Gen Genet. 261(2):259–266.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Maughan PJ, Chaney L, Lightfoot DJ, Cox BJ, Tester M, Jellen EN, Jarvis DE. 2019. Mitochondrial and chloroplast genomes provide insights into the evolutionary origins of quinoa (Chenopodium quinoa Willd.). Sci Rep. 9(1):185.
- Min J, Kim Y, Xi H, Jang T, Kim G, Park J, Park J-H. 2019. The complete chloroplast genome of a new candidate cultivar, Sang Jae, of Abeliophyllum distichum Nakai (Oleaceae): initial step of A. distichum intraspecies variations atlas. Mitochondrial DNA Part B. 4(2):3716–3718. doi:10.1080/23802359.2019.1679678.
- Min J, Kim Y, Xi H, Heo KI, Park J. 2019. The complete chloroplast genome of coffee tree, Coffea arabica L. ‘Typica’ (Rubiaceae). Mitochondrial DNA Part B. 4(2):2240–2241.
- Min J, Park J, Kim Y, Kwon W. 2019. The complete chloroplast genome of Artemisia fukudo Makino (Asteraceae): providing insight of intraspecies variations. Mitochondrial DNA Part B. 4(1):1510–1512.
- O’brien TG, Kinnaird MF. 2003. Caffeine and conservation. Washington, D.C: American Association for the Advancement of Science.
- Oh SH, Suh HJ, Park J, Kim Y, Kim S. 2019. The complete chloroplast genome sequence of Goodyera schlechtendaliana in Korea (Orchidaceae). Mitochondrial DNA Part B. 4(2):2692–2693.
- Park J, Min J, Kim Y, Xi H, Kwon W, Jang T, Kim G, Park J-H. 2019. The complete chloroplast genome of a new candidate cultivar, Dae Ryun, of Abeliophyllum distichum Nakai (Oleaceae). Mitochondrial DNA Part B. 4(2):3713–3715. doi:10.1080/23802359.2019.1679676.
- Park J, Kim Y. 2019. The second complete chloroplast genome of Dysphania pumilio (R. Br.) mosyakin and clemants (Amranthaceae): intraspecies variation of invasive weeds. Mitochondrial DNA Part B. 4(1):1428–1429.
- Park J, Kim Y, Kwon W, Nam S, Song M. 2019. The second complete chloroplast genome sequence of Nymphaea alba L. (Nymphaeaceae) to investigate inner-species variations. Mitochondrial DNA Part B. 4(1):1014–1015.
- Park J, Kim Y, Kwon W, Xi H, Kwon M. 2019. The complete chloroplast genome of tulip tree, Liriodendron tulifipera L.(Magnoliaceae): investigation of intra-species chloroplast variations. Mitochondrial DNA Part B. 4(2):2523–2524.
- Park J, Kim Y, Lee K. 2019. The complete chloroplast genome of Korean mock strawberry, Duchesnea chrysantha (Zoll. and Moritzi) Miq.(Rosoideae). Mitochondrial DNA Part B. 4(1):864–865.
- Park J, Kim Y, Xi H. 2019a. The complete chloroplast genome of aniseed tree, Illicium anisatum L. (Schisandraceae). Mitochondrial DNA Part B. 4(1):1023–1024.
- Park J, Kim Y, Xi H. 2019b. The complete chloroplast genome sequence of male individual of Korean endemic willow, Salix koriyanagi Kimura (Salicaceae). Mitochondrial DNA Part B. 4(1):1619–1621.
- Park J, Kim Y, Xi H, Heo KI. 2019a. The complete chloroplast genome of coffee tree, Coffea arabica L. ‘Blue Mountain’ (Rubiaceae). Mitochondrial DNA Part B. 4(2):2436–2437.
- Park J, Kim Y, Xi H, Heo KI. 2019b. The complete chloroplast genome of ornamental coffee tree. Coffea arabica L. (Rubiaceae). Mitochondrial DNA Part B. 4(1):1059–1060.
- Park J, Kim Y, Xi H, Jang T, Park JH. 2019. The complete chloroplast genome of Abeliophyllum distichum Nakai (Oleaceae), cultivar Ok Hwang 1ho: insights of cultivar specific variations of A. distichum. Mitochondrial DNA Part B. 4(1):1640–1642.
- Park J, Kim Y, Xi H, Kwon W, Kwon M. 2019. The complete chloroplast and mitochondrial genomes of Hyunsasi tree, Populus alba x Populus glandulosa (Salicaceae). Mitochondrial DNA Part B. 4(2):2521–2522.
- Park J, Kim Y, Xi H, Nho M, Woo J, Seo Y. 2019. The complete chloroplast genome of high production individual tree of Coffea arabica L. (Rubiaceae). Mitochondrial DNA Part B. 4(1):1541–1542.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019a. The complete chloroplast genome of common camellia tree in Jeju island, Korea, Camellia japonica L. (Theaceae): intraspecies variations on common camellia chloroplast genomes. Mitochondrial DNA Part B. 4(1):1292–1293.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019b. The complete chloroplast genome of common camellia tree, Camellia japonica L. (Theaceae), adapted to cold environment in Korea. Mitochondrial DNA Part B. 4(1):1038–1040.
- Park J, Xi H, Kim Y, Heo KI, Nho M, Woo J, Seo Y, Yang JH. 2019. The complete chloroplast genome of cold hardiness individual of Coffea arabica L. (Rubiaceae). Mitochondrial DNA Part B. 4(1):1083–1084.
- Samson N, Bausher MG, Lee SB, Jansen RK, Daniell H. 2007. The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnology J. 5(2):339–353.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(Suppl 14):S2.
- Zhu Q, Gao P, Liu S, Amanullah S, Luan F. 2016. Comparative analysis of single nucleotide polymorphisms in the nuclear, chloroplast, and mitochondrial genomes in identification of phylogenetic association among seven melon (Cucumis melo L.) cultivars. Breed Sci. 66(5):711–719.
