Abstract
Selaginella tamariscina (Beauv.) Spring distributed in East Asia region including Korea used as traditional medicine. We completed chloroplast genome of Korean S. tamariscina. Its length is 126,368 bp and has four subregions: 53,213 bp of large single-copy and 47,560 bp of small single-copy regions are separated by 12,796 bp of direct repeat regions including 80 genes (61 protein-coding genes, 8 rRNAs, and 9 tRNAs). Phylogenetic trees show that Korean S. tamariscina is clustered with that of China species and 1,213 single nucleotide polymorphisms and 1,641 insertions and deletions are identified against Chinese S. tamariscina.
Selaginella tamariscina (Beauv.) Spring (Selaginellaceae) possesses extreme drought tolerance (Giarola et al. Citation2017), distributing in Russia (Siberia), China, Korea, Japan, Philippines, Taiwan, northern Thailand, and India. It has long been used as a beneficial traditional medicine in Korea. Since it contains Selaginellin (Cheng et al. Citation2008), flavonoids (Zheng, Zhang, et al. Citation2011), and sumaflavone (Yang et al. Citation2006), it also presented anti-cancer (Yang et al. Citation2007), inhibition of immediate allergic reactions (Dai et al. Citation2005), and antihyperglycemic activities (Zheng, Li, et al. Citation2011).
Total DNA of S. tamariscina collected in Wolsong-ri, Jijeong-myeon, Wonju-si, Korea (Voucher in InfoBoss Cyber Herbarium (IN); K. Lee, IB-01028; 37.442160 N, 127.824103E) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome was sequenced using HiSeqX at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and SOAPGapCloser 1.12 (Zhao et al. Citation2011) and confirmed by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for chloroplast genome annotation based on the Chinese S. tamariscina chloroplast (NC_041646; Xu et al. Citation2018).
The chloroplast genome of Korean S. tamariscina (GenBank accession is MN894555) is 126,368 bp (GC ratio: 54.0%) and has four subregions: 53,213 bp of large single copy (53.4%) and 47,560 bp of small single copy (54.0%) regions are separated by 12,796 bp of direct repeat regions (55.3%). It contains 80 genes (61 protein-coding genes, 8 rRNAs, 9 tRNAs, and 2 pseudogenes); 7 genes (2 protein-coding genes, 4 rRNAs, and 1 tRNA) are duplicated in direct repeat regions. One pseudogene, chlN, is truncated by one SNP at 103,153 bp on chloroplast genome, which occurred in common ancestor of angiosperm (Jansen et al. Citation2007).
Alignment of two S. tamariscina chloroplast genomes presents 1,213 single nucleotide polymorphisms (SNPs) and 1,641 insertions and deletions (INDELs) are identified, higher than those of Selaginella uncinate between Japan (Tsuji et al. Citation2007) and China (Zhang et al. Citation2019; 525 SNPs and 602 INDELs). It is similar level to Goodyera schlechtendaliana (711 to 2,889 variations; Oh et al. Citation2019a, Citation2019b) and higher than Pyrus ussuriensis (121 SNPs and 781 INDELs; Cho et al. Citation2019), Camellia japonica (78 SNPs and 643 INDELs; Park et al. Citation2019), and Marchantia polymorpha (69 SNPs and 660 INDELs; Kwon et al. Citation2019). Proportion of variations on the chloroplast genome of S. tamariscina is 2.26%, which is between G. schlechtendaliana (1.95%; Oh et al. Citation2019a, Citation2019b) and Gastrodia electa (3.04%; Park et al. Citation2020).
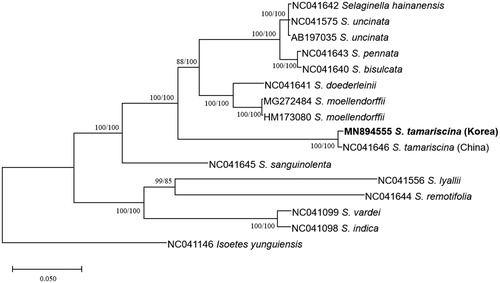
Fifteen chloroplast genomes of Selaginella including two S. tamariscina chloroplasts and one outgroup species, Isoetes yunguiensis (Zhou et al. Citation2016), were used for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood phylogenic trees (bootstrap repeat is 1,000) using MEGA X (Kumar et al. Citation2018) after concatenating alignments of 44 conserved genes by MAFFT 7.450 (Katoh and Standley Citation2013) under the environment of Plant Chloroplast Database (http://www.cp-genome.net/). Phylogenetic trees show that Korean S. tamariscina is clustered with that of China ().
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic tree of 44 conserved genes from 15 Selagenella and outgroup species chloroplast genomes: Selaginella tamariscina (MN894555 in this study, NC_041646), S. bisulcata (NC_041640), S. doederleinii (NC_041641), S. hainanensis (NC_041642), S. indica (NC_041098), S. lyallii (NC_041556), two S. moellendorffii (HM173080 and MG272484), S. pennata (NC_041643), S. remotifolia (NC_041644), S. sanguinolenta (NC_041645), two S. uncinata (NC_041575 and AB197035), S. vardei (NC_041099), and outgroup species, Isoetes yunguiensis (NC_041146). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cheng X-L, Ma S-C, Yu J-D, Yang S-Y, Xiao X-Y, Hu J-Y, Lu Y, Shaw P-C, But PP-H, Lin R-C. 2008. Selaginellin A and B, two novel natural pigments isolated from Selaginella tamariscina. Chem Pharm Bull. 56(7):982–984.
- Cho MS, Kim Y, Kim SC, Park J. 2019. The complete chloroplast genome of Korean Pyrus ussuriensis Maxim.(Rosaceae): providing genetic background of two types of P. ussuriensis. Mitochondrial DNA Part B. 4(2):2424–2425.
- Dai Y, But PP-H, Chu L-M, Chan Y-P. 2005. Inhibitory effects of Selaginella tamariscina on immediate allergic reactions. Am J Chin Med. 33(6):957–966.
- Giarola V, Hou Q, Bartels D. 2017. Angiosperm plant desiccation tolerance: hints from transcriptomics and genome sequencing. Trend Plant Sci. 22(8):705–717.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci. 104(49):19369–19374.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kwon W, Kim Y, Park J. 2019. The complete mitochondrial genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: inverted repeats on mitochondrial genome between Korean and Japanese isolates. Mitochondr DNA B. 4(1):769–770. DOI:10.1080/23802359.2019.1565975
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. :13033997.
- Li H, 1000 Genome Project Data Processing Subgroup, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019a. The complete chloroplast genome sequence of a morphotype of Goodyera schlechtendaliana (Orchidaceae) with the column appendages. Mitochondr DNA B. 4(1):626–627.
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019b. The complete chloroplast genome sequence of Goodyera schlechtendaliana in Korea (Orchidaceae). Mitochondr DNA B. 4(2):2692–2693.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019. The complete chloroplast genome of common camellia tree, Camellia japonica L.(Theaceae), adapted to cold environment in Korea. Mitochondr DNA B. 4(1):1038–1040.
- Park J, Suh Y, Kim S. 2020. A complete chloroplast genome sequence of Gastrodia elata (Orchidaceae) represents high sequence variation in the species. DOI:10.1080/23802359.2019.1710588
- Tsuji S, Ueda K, Nishiyama T, Hasebe M, Yoshikawa S, Konagaya A, Nishiuchi T, Yamaguchi K. 2007. The chloroplast genome from a lycophyte (microphyllophyte), Selaginella uncinata, has a unique inversion, transpositions and many gene losses. J Plant Res. 120(2):281–290.
- Xu Z, Xin T, Bartels D, Li Y, Gu W, Yao H, Liu S, Yu H, Pu X, Zhou J, et al. 2018. Genome analysis of the ancient tracheophyte Selaginella tamariscina reveals evolutionary features relevant to the acquisition of desiccation tolerance. Mol Plant. 11(7):983–994.
- Yang JW, Pokharel YR, Kim M-R, Woo E-R, Choi HK, Kang KW. 2006. Inhibition of inducible nitric oxide synthase by sumaflavone isolated from Selaginella tamariscina. J Ethnopharmacol. 105(1–2):107–113.
- Yang SF, Chu SC, Liu SJ, Chen YC, Chang YZ, Hsieh YS. 2007. Antimetastatic activities of Selaginella tamariscina (Beauv.) on lung cancer cells in vitro and in vivo. J Ethnopharmacol. 110(3):483–489.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang H-R, Xiang Q-P, Zhang X-C. 2019. The unique evolutionary trajectory and dynamic conformations of DR and IR/DR-coexisting plastomes of the early vascular plant Selaginellaceae (Lycophyte). Genome Biol Evol. 11(4):1258–1274.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(Suppl 14):S2.
- Zheng XK, Li YJ, Zhang L, Feng WS, Zhang X. 2011. Antihyperglycemic activity of Selaginella tamariscina (Beauv.) spring. J Ethnopharmacol. 133(2):531–537.
- Zheng XK, Zhang L, Wang WW, Wu YW, Zhang QB, Feng WS. 2011. Anti-diabetic activity and potential mechanism of total flavonoids of Selaginella tamariscina (Beauv.) Spring in rats induced by high fat diet and low dose STZ. J Ethnopharmacol. 137(1):662–668.
- Zhou X-M, Rothfels C J, Zhang L, He Z-R, Le Péchon T, He H, Lu N T, Knapp R, Lorence D, He X-J, et al. 2016. A large‐scale phylogeny of the lycophyte genus Selaginella (Selaginellaceae: Lycopodiopsida) based on plastid and nuclear loci. Cladistics. 32(4):360–389.
