Abstract
Cirsium shansiense is a common species in China, which can be used for traditional Chinese medicine. The chloroplast (cp) genome of C. shansiense exhibited a typical quadripartite cycle with 152,396 bp, including a pair of inverted repeat regions (IRa and IRb, 25,072 bp each), a large single-copy region (LSC, 83,517 bp), and a small single-copy region (SSC, 18,735 bp). One-hundred-thirty-three unique genes were assembled in the cp genome, including 88 protein genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on 17 cp genomes of related species of Asteraceae showed that C. shansiense was closely related to Cirsium vulgare. This study provides important information for identification and conservation of species of Cirsium spp.
Cirsium shansiense is a widespread herb species in China. It is mainly distributed in mountain forests, wetlands, and roadside (Editorial Committee of Flora of China, Chinese Academy of Sciences Citation1987). There are many variations of population patterns (Shih Citation1984), hence, it is often wrongly identified as other species of Cirsium spp. (e.g. Cirsium chinense, Shih Citation1984). Moreover, C. shansiense is also a kind of traditional Chinese medicine source species, which is mainly used to treat bleeding, infectious diseases, and hypertension (Tao et al. Citation2012). However, studies on C. shansiense based on molecular biology have not been reported. Here, the chloroplast (cp) genome of C. shansiense was assembled. Also, a phylogenetic analysis of C. shansiense was performed. These studies will provide more evidence for the identification and application of C. shansiense.
The fresh leaves (about 5 g) of a single individual of C. shansiense were sampled from Xifeng district, Qingyang city, Gansu Province, China (107°34′5′′E, 35°42′51′′N). There is a planted forest habitat on Loess Plateau. A voucher specimen (WNU52012) was deposited at the herbarium, in the School of Life Sciences, Northwest University. Genomic DNA was extracted from the fresh leaves by a modified CTAB method (Doyle Citation1987). Total DNA was used for library construction and then sequencing on Illumina HiSeq 4000. Illumina reads were mapped to the assembly using Bowtie2 v2.1.0 (Langmead and Salzberg Citation2012) and then finishing the assembly by MITObim v1.8 (Hahn et al. Citation2013) with the reference sequence of Cirsium rhinoceros (GenBank: NC_044423). The complete cp genome was submitted to GenBank (accession number: MN871982).
The complete cp genome length of C. shansiense was 152,396 bp, with a large single-copy region (LSC, 83,517 bp), a small single-copy region (SSC,18,735 bp), and two inverted repeats (IRa and IRb, 25,072 bp each). The overall GC content of C. shansiense cp genome was 37.7%. The cp genome of C. shansiense contained 133 genes, including 88 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. A total of 30 genes were duplicated in the IR regions, including 9 protein-coding genes, 13 tRNA genes, and 8 rRNA genes. The ycf1 gene was located at the SSC and IRa junction.
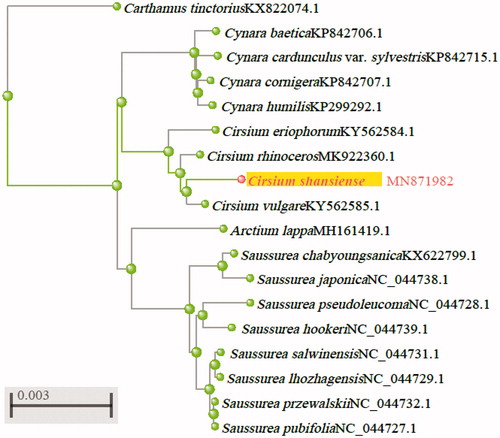
To investigate the phylogenetic relationships with closely related species, the neighbor-joining(NJ) phylogenetic tree was constructed by BLAST tools on NCBI (https://blast.ncbi.nlm.nih.gov) based on 17 complete cp genomes (download from Genbank, ) of Asteraceae species. A total of 1000 bootstrap replicates were computed to confirm the stability of each tree node. The tree showed a close relationship between C. shansiense and Cirsium vulgare. The complete cp genome sequence of C. shansiense can provide more information for the identification and conservation of species of Cirsium spp.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anton B, Sergey N, Dmitry A, Gurevich AA, Mikhail D, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Editorial Committee of Flora of China, Chinese Academy of Sciences. 1987. Flora of China. Beijing (China): Science Press; p. 122–123.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-abaiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Meth. 9(4):357–359.
- Shih C. 1984. Notulae de Plantis Tribus Cynarearum Familiae Compositarum Sinicae (II)(Cont.). J Syst Evol. 22(6):445–455.
- Tao M, Luo Q, Huang Y. 2012. Infrared and thermal analysis identification of cirsium shansiense petrak and potentilla discolor bunge. Med Plant. 10:49–50.