Abstract
The complete mitogenome of Jaydia carinatus was sequenced, that was a closed double-stranded circular molecule of 16,455 bp. We analyzed the main features of this sequence in terms of the genome organization, gene arrangement and codon usage. The overall base composition included A(27.6%), T(25.7%), C(29.6%) and G(17.1%). Moreover, the 13 protein-coding genes (PCGs) encoded 3821 amino acids in total, 12 of which used the initiation codon ATG except COI used GTG; most of them had a complete stop codon, whereas Cytb ended with the incomplete stop codon T. The phylogenetic tree was conducted to provide relationship within Kurtiformes based on the Neighbor Joining method and the NJ tree demonstrated this species had a closest relationship with Jaydia lineata, which could be a useful basis for propagation of this species.
Jaydia carinatus was known mainly from Western Pacific, Japan and the China seas, sometimes appears in northwestern Australia (Allen and Swainston Citation1988). The young are rather rare, while adults are usually caught by trawls and used as feed for farmed fish. As an important group of minor commercial fishes in fishing wharfs of China (Masuda Citation1984). We described the characterization of complete mitochondrial genome of J. carinatus and explored its phylogenetic relationship within Kurtiformes, which were expected to facilitate future studies on taxonomic resolution, population genetic structure and phylogenetic relationships.
Specimen of J. carinatus was sampled by commercial bottom trawling in the East China Sea (30°06′59″N; 123°29′50″E) and stored in Marine Fisheries Research Institute of Zhejiang Province with accession number 20190909JC.
The complete mitochondrial genome of J. carinatus was a closed double stranded circular molecule of 16,455 nucleotides (GenBank Accession Number: MN937193), consisting of 13 PCGs, 22 tRNA genes, two rRNA genes, one replication origin and a control region, that was similar to the typical mitogenome of other vertebrates (Miya et al. Citation2003; Zhu et al. Citation2018). The overall base composition was 27.6%, 25.7%, 29.6% and 17.1% for A, T, C and G respectively, with a slight AT bias 53.3%. The 13 PCGs genes encoded 3821 amino acids in total, and all the PCGs used the initiation codon ATG except COI used GTG (Miya and Nishida Citation2000). Most of them used TAA or TAG as the stop codon, three PCGs (COII, ND4 and ND6) use AGA as the stop codon, while one PCG used an incomplete stop codon T, these incomplete termination codons were presumably completed as TAA via post-transcriptional polyadenylation (Ojala et al. Citation1981). The 12S and 16S were 956 bp and 1684 bp respectively. The tRNA genes were generated using the program tRNAs-can-SE, all of them could fold into a typical cloverleaf structure except tRNA-Ser (AGY), devoiding of a dihydrouridine arm. As in other vertebrates, two typical non-coding regions were detected in the J. carinatus mitogenome, the OL was located in a cluster of five tRNA genes (WANCY), which had the potential to fold into a stable stem-loop secondary structure (Lynch Citation1997), with a stem formed by 13 paired nucleotides and a loop of 13 nucleotides (Petrillo et al. Citation2006); the CR, with 858 bp, was situated between tRNA-Pro and tRNA-Phe, and the core sequence of the terminal-associated sequence (ACATATATG) was identified in the CR of J. carinatus, which was identical to that in other teleostean mitogenomes (Zhang et al. Citation2013).
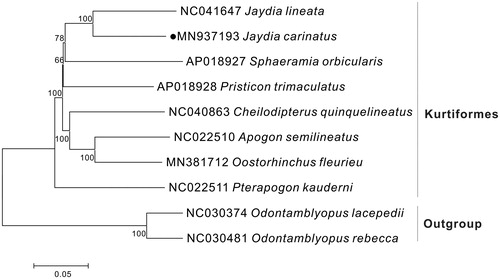
To explore the phylogenetic position of this J. carinatus, we constructed a phylogenetic tree based on the NJ analysis of 12 PCGs encoded by the heavy strand of eight Kurtiformes species. The result of present study supports J. carinatus has a closest relationship with J. lineata (), that would contribute to the understanding of the phylogeny of Kurtiformes.
Figure 1. Neighbor Joining (NJ) tree of 8 Kurtiformes species based on 12 PCGs. The bootstrap values are based on 1000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labeled with a black spot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Allen GR, Swainston R. 1988. The marine fishes of north-western Australia: a field guide for anglers and divers. Perth (Australia): Western Australian Museum.
- Lynch M. 1997. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol Biol Evol. 14:914.
- Masuda H. 1984. The fishes of the Japanese Archipelago. Tokyo: Tokai University Press.
- Miya M, Nishida M. 2000. Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Phylogenet Evol. 17:437–455.
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26:121–138.
- Ojala D, Montoya J, Attardi G. 1981. TRNA punctuation model of RNA processing in human mitochondrial. Nature. 290(5806):470–474.
- Petrillo M, Silvestro G, Nocera P.P.D, Boccia A, Paolella G. 2006. Stem-loop structures in prokaryotic genomes. BMC Genom. 7(1):170.
- Zhang H, Zhang Y, Zhang X, Song N, Gao T. 2013. Special structure of mitochondrial DNA control region and phylogenetic relationship among individuals of the black rockfish, Sebastes schlegelii. Mitochondrial DNA. 24(2):151–157.
- Zhu K, Lü Z, Liu L, Gong L, Liu B. 2018. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3(1):301–302.
