Abstract
We report the complete mitogenome for the California grunion (Leuresthes tenuis), a beach-spawning silverside fish endemic to eastern Pacific coastal waters of California, USA and Baja California, Mexico. Approximately, 28,428 short reads from a shotgun gDNA library were mapped to a reference mitogenome of Menidia menidia. The resulting L. tenuis mitogenome is 16,719 bases in length. We further provide a Bayesian phylogenetic tree showing relationships among L. tenuis and other Atheriniform fishes.
Keywords:
The California grunion Leuresthes tenuis (Atheriniformes) historically ranges from south of Point Conception, California, USA, to Bahía Magdalena, Baja California Sur, Mexico (Miller and Lea Citation1972), with reports over the last decade north to San Francisco Bay and Tomales Bay (Yoklavich et al. Citation2002, Jahn Citation2004, Roberts et al. Citation2007, Martin et al. Citation2013). California grunion and its sister species, the Gulf grunion L. sardina, are the only fish species known to spawn on sandy beaches and deposit fertilized eggs in the sand to incubate terrestrially (Thompson Citation1919, Walker Citation1952, Martin Citation2015). Here, we provide the first complete mitogenome of L. tenuis (GenBank accession: MN181432).
Genomic DNA was extracted from a single larval L. tenuis individual hatched from gametes stripped from adults collected at Doheny State Beach, Dana Point, CA, USA, (33.461667, −117.687222) on 23 April 2016, using the QIAgen DNeasy Tissue Kit (Valencia, CA, USA) and used to construct a next-generation sequencing library (Illumina HiSeq2500) for a separate molecular marker development project. Specimen DNA is archived at the Natural History Museum of Los Angeles County (NHM) under accession number LACM:T-001274. Over 4.7 million 250-PE reads were processed for quality in Trimmomatic V0.36 (Bolger et al. Citation2014). The remaining 3.5 million reads were mapped to a Menidia menidia mitogenome (GenBank no: AB370893) in Geneious 10.2.3 (Kearse et al. Citation2012), and all polymorphic sites were inspected for sufficient coverage (50–438x). Annotations were transferred from M. menidia, and the resulting full L. tenuis mitogenome (GenBank no: MN181432, NC_044649) was then aligned with available Atheriniform mitogenomes (GenBank accessions in ) using MAFFT 7.0 (Katoh et al. Citation2019). A Bayesian phylogram was constructed using MrBayes 3.2.2 (Ronquist and Huelsenbeck Citation2003, Ronquist et al. Citation2012).
Figure 1. Bayesian phylogram of Atheriniform mitogenomes. Node support values are Bayesian posterior probabilities.
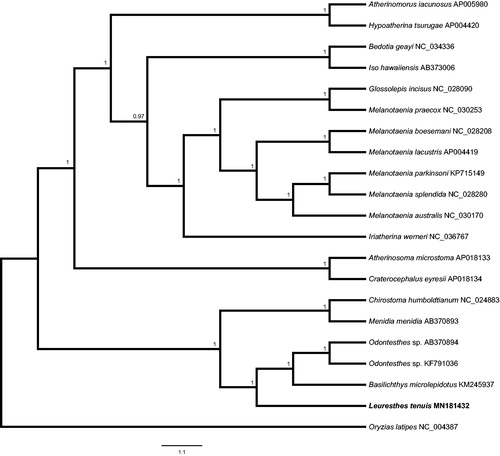
The 16,719 base pair L. tenuis mitogenome possesses 13 protein-coding genes, 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (rRNA), and a D-loop (control region), approximately 200–300 bp longer than other Atheriniform species. Tree reconstruction places L. tenuis in a group with B. microlepidotus and Odontesthes sp. () with M. menidia and Chirostoma humboldtianum comprising a sister group. The resulting Atheriniform mitogenomes phylogeny yields an incomplete phylogenetic reconstruction, not fully reflective of published relationships among Atheriniform fishes (Dyer and Chernoff Citation1996, Parenti Citation2005, Campanella et al. Citation2015). Future work should focus on comparisons among mitogenomes of the two Leuresthes species, L. tenuis, and L. sardina and other close relatives, to further resolve relationships among these eastern Pacific endemic species.
Acknowledgments
Thanks to William Lundt for assistance with archival of the L. tenuis specimen DNA at NHM. Adult grunion used to generate the specimen in this work were collected under CDFW scientific collecting permit #SC 3211 to KAD (and CSUF IACUC-approved protocol #17-R-06).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Campanella D, Hughes LC, Unmack PJ, Bloom DD, Piller KR, Ortí G. 2015. Multi-locus fossil-calibrated phylogeny of Atheriniformes (Teleostei, Ovalentaria). Mol Phylogenetics Evol. 86:8–23.
- Dyer BS, Chernoff B. 1996. Phylogenetic relationships among Atheriniform fishes (Teleostei: Atherinomorpha). Zool J Linn Soc. 117(1):1–69.
- Jahn A. 2004. Summertime distribution of three species of atherinopsid fishes in east-central San Francisco Bay. Bull South Calif Acad Sci. 103:34–35.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Kearse M, Moir R, Wilson A, Stone-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markovitz S, Duran C, et al. 2012. Geneius basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Martin KLM, Hieb KA, Roberts DA. 2013. A California icon surfs north: local ecotype of California grunion Leuresthes tenuis (Atherinopsidae) revealed by multiple approaches during temporary habitat expansion into San Francisco Bay. Copeia. 2013(4):729–739.
- Martin K. 2015. Beach-spawning fishes: reproduction in an endangered ecosystem. Boca Raton, Florida: CRC Press; p. 219.
- Miller DJ, Lea RN. 1972. Guide to the coastal marine fishes of California. Fish B Calif Fish Game. 157:1–235.
- Parenti LR. 2005. The phylogeny of Atherinomorphs: evolution of a novel fish reproductive system. In: Uribe MC, Grier HJ, editors. Viviparous fishes. Homestead, Florida: New Life Publications; p. 13–30, 603.
- Roberts D, Lea RN, Martin K. 2007. First record of the occurrence of the California grunion, Leuresthes tenuis, in Tomales Bay, California: a northern extension of the species. Calif Fish Game. 93:107–110.
- Thompson WF. 1919. The spawning of the grunion (Leuresthes tenuis). Fish B Calif Fish Game. 3:1–29.
- Walker BW. 1952. A guide to the grunion. Calif Fish Game. 38:409–420.
- Yoklavich MA, Cailliet GM, Oxman DS, Barry JP, Lindquist DC. 2002. Fishes. In: Caffrey J, Brown M, Tyler WB, Silberstein M, editors. Changes in a California estuary: a profile of Elkhorn Slough. Moss Landing (CA): Elkhorn Slough Foundation; p. 163–185.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MRBAYES 3.2: efficient Bayesian phylogenetic inference and model selection across a large model space. Syst Biol. 61(3):539–542.
