Abstract
Hosta ensata (Asparagaceae) is a perennial plant with great value for its decorative leaves and colorful flowers, which is massively cultivated in parks and gardens for ornamental purposes. Here, the complete chloroplast genome of the H. ensata has been constructed from the Illumina sequencing data. The circular cp genome is 156,712 bp in size and comprises of a pair of inverted repeat (IR) regions of 24,646 bp each, a large single-copy (LSC) region of 85,284 bp, and a small single-copy (SSC) region of 22,136 bp. The total GC content is 37.8%, while the corresponding values of the LSC, SSC, and IR regions are 35.9, 33.1, and 43.3%, respectively. The chloroplast genome contains 131 genes, including 85 protein-coding genes, 8 ribosomal RNA genes, and 38 transfer RNA genes. The maximum-likelihood phylogenetic analysis showed a strong sister relationship with Hosta yingeri and Hosta capitata in Hosta. Our findings provide a foundation for further investigation of chloroplast genome evolution in ornamental plant Hosta.
Hosta ensata (Asparagaceae) native to Southern of Jilin, Liaoning province in Northeast of China, is a perennial plant with great value for its decorative leaves and colorful flowers. It has been massively cultivated in parks and gardens for ornamental purposes (Liu et al. Citation2019). Besides, the characteristics of cold resistance, high reproduction coefficient, and simple field management makes H. ensata a high-quality ground cover planted widely. In spite of its horticultural and economic importance, there are a few genetic and genomic studies for breeding of this plant. Therefore, we reported the complete chloroplast genome (cp) of H. ensata based on Illumina sequencing data, which would be helpful for its evolution and genetics research.
Fresh leaves of H. ensata were collected from Baishi Mountain in Jilin city, Jinlin province of China (126°46′43″E, 43°24′47″N), for total genomic DNA extraction. The voucher specimen was preserved at the Herbarium of Jilin Agriculture Universtiy (accession number YZDB1910328). High-throughput DNA sequencing was conducted on the Illumina HiSeq 2500 Sequencing System (Illumina, CA, USA), and sequenced by Genesky Biotechnology (Shanghai, China). We assembled the cp genome using CLC Genomics Workbench v7.5 (CLC Bio, Aarhus, Denmark) as stated previously (Zuo et al. Citation2017). A subset of 36.57 M trimmed reads were used for reconstructing the chloroplast genome by NOVOPlasty (Dierckxsens et al. Citation2017), with that of its congener Hosta minor (GenBank: NC_035999.1) as the initial reference genome. The physical map of the new chloroplast genome was generated using MITObim v1.8 (Hahn et al. Citation2013). Finally, the validated complete chloroplast genome sequence was submitted to GenBank with accession number MN901630.
The complete chloroplast genome of H. ensata is 156,712 bp in size with high coverage (mean 339.2×), containing a pair of inverted repeat (IR) regions of 24,646 bp each, separated by a large single-copy (LSC) region of 85,284 bp, and a small single-copy (SSC) region of 22,136 bp. The total GC content is 37.8%, while the corresponding values of the LSC, SSC, and IR region are 35.9, 33.1, and 43.3%, respectively. This chloroplast genome harbors 131 functional genes, including 85 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Among them, 53 are involved in photosynthesis and 68 genes are involved in self-replication. Seven protein-coding genes, rpl2, rpl23, rps7, rps12, rps19, ndhB, and ycf2 were duplicated genes which is located in IR regions. Besides, 18 genes contain one intron, while chlp, rps12, and ycf3 harbor two introns. Summarily, the H. ensata cp genome is structurally similar to previously published ones (Lim et al. Citation2017; Jang et al. Citation2018).
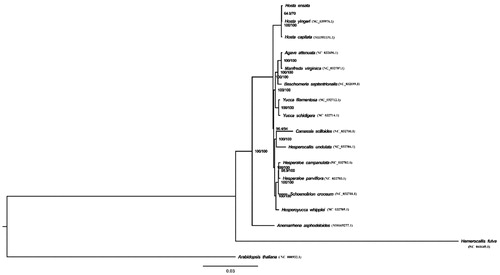
The phylogenetic tree was generated based on the complete cp genome of H. ensata and other 16 species (). The alignment was conducted using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was built using the maximum likelihood (ML) method. The results showed that H. ensata was closely related to Hosta yingeri and Hosta capitata in Hosta. Our findings provide a foundation for further investigation of chloroplast genome evolution in ornamental plant Hosta.
Figure 1. The phylogenetic tree was constructed using chloroplast genome sequences of 17 species within the Asparagaceae family and Arabidopsis thaliana as an outgroup based on the maximum-likelihood analysis using 500 bootstrap replicates. Chloroplast genome sequences used for this tree are Anemarrhena asphodeloides, NC_032698.1; Anemarrhena asphodeloides, MH669277.1; Arabidopsis thaliana, NC_000932.1; Beschorneria septentrionalis, NC_032699.1; Camassia scilloides, NC_032700.1; Hemerocallis fulva, NC_041649.1; Hesperaloe parviflora, NC_032703.1; Hesperaloe campanulata, NC_032702.1; Hesperocallis undulata, NC_032704.1; Hesperoyucca whipplei, NC_032705.1; Hosta capitata, MH581151.1; Hosta yingeri, NC_039976.1; Hemerocallis fulva, NC_041649.1; Manfreda virginica, NC_032707.1; Schoenolirion croceum, NC_032710.1, Yucca filamentosa, NC_032712.1; Yucca schidigera, NC_032714.1.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Jang Y, Park JY, Kang SJ, Park SH, Shim H, Lee TJ, Kang JH, Sung SH, Yang TJ. 2018. The complete chloroplast genome sequence of Hosta capitata (Koidz.) Nakai (Asparagaceae). Mitochondr DNA B. 3(2):1052–1053.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lim CE, Kim S, Lee HO, Ryu S-A, Lee B-Y. 2017. The complete chloroplast genome of a Korean endemic ornamental plant Hosta yingeri SB Jones (Asparagaceae). Mitochondrial DNA Part B. 2(2):800–801.
- Liu ZX, Dong AR, Jia HQ, Zhou Y, Liu XF, Zhang XY. 2019. First Report of Pseudomonas syringae pv. syringae causing leaf blight on Hosta ventricosa in China. Plant Disease. 103(8):2123.
- Zuo LH, Shang AQ, Zhang S, Yu XY, Ren YC, Yang MS, Wang JM. 2017. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: genome comparative and taxonomic position analysis. PLoS One. 12(2):e0171264.
