Abstract
The complete mitochondrial genome of Putian Loquat longicorn (Anoplophora chinensis) was determined in this study. Its mitochondrial genome (15,871 bp) contains 13 protein-coding genes, 22 tRNA genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), and one large non-coding region (D-loop region). Nine protein-coding genes and fourteen tRNA genes are encoded on the H strand, and the other four protein-coding genes, two rRNA genes, and eight tRNA genes are encoded on the L strand. The arrangement of genes is identical to all know Longhorn beetle mitochondrial genomes. The complete mitochondrial genome of the A. chinensis provides an important data set for further study on its classification.
Cerambycidae is a large family in the Coleoptera order. Most of the species in the Cerambycidae family are fruiter pests. To date, more than 25,000 species of Cerambycidae have been described worldwide (Wang et al. Citation2012), and more than 3100 Cerambycidae in China (Wang and Hua Citation2009). Despite the large taxonomic diversity within this family, information about the Cerambycidae genome is still limited. Putian is the main producing area of Chinese Loquat and has the reputation of the hometown of Loquat. Putian Loquat longicorn is one of the main insects that cause the decrease of loquat yield and the death of loquat tree in Putian aera. Here, the complete mitochondrial genome of Putian Loquat longicorn (Anoplophora chinensis) was sequenced and characterized in detail. The specimens were collected from Putian (25°28′N, 118°40′E), Fujian Province, China in August 2019. The specimen of Putian Loquat longicorn, named as AchinPut-01, was stored in College of Life Sciences, Linyi University, Linyi, China. Total genomic DNA was extracted from Putian Loquat longicorn (A. chinensis) muscle according to Liu et al. (Citation2016), Li et al. (Citation2019), and Yao et al. (Citation2019). Then, the complete mitochondrial genome was sequenced using a shotgun approach and assembly. Thereafter, DNA sequence was analyzed using MEGA 7 (Kumar et al. Citation2016) and protein-coding genes were analyzed using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) using the invertebrate mitochondrial code. The tRNA genes were identified by ARWEN (Laslett and Canbäck Citation2008) and tRNA-scan SE (Lowe and Eddy Citation1997).
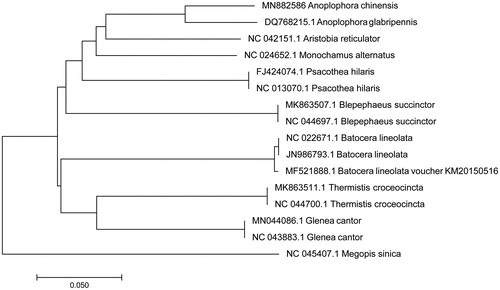
The mitochondrial genome of Putian Loquat longicorn (Accession number MN882586) was 15,871 in length, of which 14,632 nucleotides are coding DNA, and 1239 nucleotides are non-coding DNA. It contains the 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and one control region. The arrangement and composition of the mitochondrial genome is also comparable to the case of other Arthropoda (Lavrov et al. Citation2000; Masta and Boore Citation2004; Choi et al. Citation2007). A phylogenetic tree was constructed based on the comparison of Putian Loquat longicorn mitochondrial genome sequences with other Cerambycidae species using Neighbour-Joining method (). There were 11 overlapping regions (total 32 bp) and 10 intergenic spacers (total 42 bp) among the genes. The total base composition of the mitochondrial genome is 39.47% A, 38.14% T, 8.81% G and 13.58% C, and an A + T (77.61%)-rich feature occurs in the Putian Loquat longicorn. To investigate the nucleotide bias, skew for a given strand was calculated as (A-T)/(A + T) or (G-C)/(G + C) (Perna and Kocher Citation1995). The AT and GC skews for the Putian Loquat longicorn mitochondrial genome were 0.017 and −0.213, respectively; this finding indicated that the strand that encoded genes contained more A and C than T and G, and this skew was evidence of codon usage bias. All 13 protein-coding genes identified in the Putian Loquat longicorn mitochondrial genome, the initiation codons of ND2, COX2, ATP8, ND3, and ND6 were ATT, the initiation codons for ATP6, COX3, and CYTB were ATG, whereas ND4L, COX1, ND5, ND4, and ND1, which began with AAA, GAA, ATA, TAT, and CTA, respectively. The termination codons for ND2, ATP8, ATP6, COX3, and ND6 genes were TAA, the termination codon for CYTB was TAG. In addition, the 7 remaining genes terminate with incomplete termination codon T—, that T— is the 5’ terminal of the adjacent gene, which presumptively formed a complete stop codon by post-transcriptional polyadenylation (Anderson et al. Citation1981). Except for four protein-coding genes (ND4, ND4L, ND1, and ND5), two rRNA genes, and eight tRNA genes (tRNA-Gln, Cys, Tyr, Phe, His, Pro, Leu, and Val), all the mitochondrial genome genes were encoded on the H strand. The 16S rRNA (1273 bp) gene and 12S rRNA (801 bp) gene were located between the D-loop and tRNA-Leu genes and separated by the tRNA-Val gene. The control region was located between 12S rRNA and tRNA-Ile with a length of 1197 bp.
Figure 1. A phylogenetic tree constructed based on the comparison of mitochondrial genome sequences of the Putian Loquat longicorn (Anoplophora chinensis) and other 15 species of Lamiinae family. Megopis sinica is used as an outgroup. GenBank accession numbers for all sequences are listed in the figure. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.
Disclosure statement
The authors report no conflicts of interest, as well as only the authors are responsible for the content and writing of the article.
Additional information
Funding
References
- Anderson S, Bankier A. T, Barrell B. G, de Bruijn M. H. L, Coulson A. R, Drouin J, Eperon I. C, Nierlich D. P, Roe B. A, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290(5806):457–465.
- Choi HE, Park SJ, Jang HK, Hwang W. 2007. Complete mitochondrial genome of a Chinese scorpion Mesobuthus martensii (Chelicerata, Scorpiones, Buthidae). DNA Sequence. 18(6):461–473.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics (Oxford, England). 24(2):172–175.
- Lavrov DV, Boore JL, Brown WM. 2000. The complete mitochondrial DNA sequence of the Horseshoe Crab Limulus polyphemus. Mol Biol Evol. 17(5):813–824.
- Li YY, Liu LX, Sui ZH, Ma C, Liu YG. 2019. The complete mitochondrial DNA sequence of Mongolian grayling Thymallus brevirostris. Mitochondrial DNA Part B. 4(1):1204–1205.
- Liu YG, Li YY, Meng W, Liu LX. 2016. The complete mitochondrial DNA sequence of Xinjiang arctic grayling Thymallus arcticus grubei. Mitochondr DNA B. 1(1):724–725.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Masta SE, Boore JL. 2004. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol. 21(5):893–902.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358.
- Wang CY, Feng Y, Chen XM. 2012. Complete sequence and gene organization of the mitochondrial genome of Batocera lineolate Chevrolat (Coleoptera: Cerambycidae). Chin Sci Bull. 57(27):3578–3585.
- Wang ZC, Hua LZ. 2009. Collect and revision of name list on Longicorn beetles in China. J Beihua Univ (Nat Sci). 10(2):159–192.
- Yao CY, Li YY, Liu LX, Ma C, Liu YG, Liu YH. 2019. The complete mitochondrial DNA sequence of Yimeng wool rabbit. Mitochondr DNA B. 4(2):3858–3859.
