Abstract
Cymbidium maguanense is a valuable orchid endemic to China, with delicate smelling, ivory white flower, and showy yellow labellum. Herein, we reported the complete chloroplast genome of C. maguanense. This chloroplast genome was 156,530 bp in length. It presented a typical structure including a large single-copy region (LSC, 85,112 bp), a small single-copy region (SSC, 17,874 bp), and two inverted repeat regions (IRs, 26,772 bp). It contained 138 genes, 37 tRNA, and 8 rRNA with an average GC content of 36.8%. Besides, the phylogenetic analysis showed that C. maguanense displayed a closer kinship to C. eburneum in the section Eburnea under subgenus Cymbidium. This chloroplast genome will provide a reference for future studies on species evolution, germplasm resources utilization, and genetic engineering of genus Cymbidium.
Cymbidium is one of the ornamental genus in Orchidaceae, including 3 subgenera and 11 sections in China (Liu et al. Citation2009). Cymbidium maguanense is an epiphytic orchid in Cymbidium Sw. sect. Eburnea under subgenus Cymbidium, distributed in southeast Yunnan at an altitude of 1000–1800 m (Liu Citation2006). It is very valuable in ornament applications with delicate smelling, ivory white flower, and showy yellow labellum (Chen Citation2011). Cymbidium maguanense is originally described by F. Y. Liu based on a cultivated plant. Its morphology is intermediate between the sympatric species C. mastersii and C. eburneum, whether it is a natural hybrid is still controversial. Morphological similarity also makes it difficult to be distinguished from related taxa in terms of species identification. The origin and classification of C. maguanense are unclear. Besides, related genetic information and relationship based on it have not been clearly formulated. Therefore, we reported the complete chloroplast genome of C. maguanense to obtain a better understanding of the relationship between C. maguanense and its related taxa, and provide a foundation to species evolution, identification, and germplasm diversity of genus Cymbidium.
Cultivated plant samples of C. maguanense were acquired from Yunnan and preserved at Fujian Agriculture and Forestry University (Voucher specimen: JL-FJ2019-11A, FAFU, 26°20′21.3″ N, 113°12′39.6″ E). DNA from fresh leaves was extracted using a modified CTAB method and sequenced on the BGI-500 platform (BGI, Wuhan, China). Approximately, 6 GB data were generated after removing adapters and unqualified reads by fastp software. Genome sequences were assembled with SPAdes 3.13.1, then annotated for genes and tRNA using Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004) and Geneious ver. 2020.0.4 (Li et al. Citation2019), to adjust the starting position by comparing with the complete chloroplast genome of C. ensifolium (NC028525).
The complete chloroplast genome of C. maguanense (GenBank accession MN885494) was 156,530 bp in length with an average GC content of 36.8%. This plastid genome presented a typical structure, including a length of 85,112 bp large single-copy (LSC) region, a 17,874 bp small single-copy (SSC) region, and two conserved 26,772 bp inverted repeat (IRs). Every part showed unequal GC contents, respectively, LSC-34.3%, SSC-29.6%, and IR-43.1%. The complete chloroplast genome contained 138 genes, 37 tRNA, and 8 rRNA.
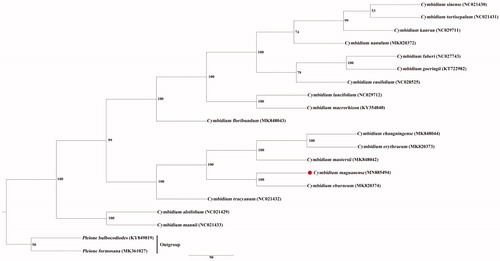
Cymbidium maguanense, C. mastersii, and C. eburneum are similar in morphological characters. The classification relationship among them is controversial (Liu et al. 2009; Liu Citation2006). To determine the phylogenetic position of C. maguanense, the complete chloroplast genome of C. maguanense was aligned with 17 accessions of Cymbidium and 2 other genera (Pleione bulbocodioides, Pleione formosana) as outgroups using HomBlocks pipeline (Bi et al. Citation2018). A molecular phylogenetic tree was constructed with the maximum parsimony (MP) in PAUP v4.0b10 (Cummings Citation2004). The result indicated that C. maguanense displayed a more close kinship to C. eburneum in the section Eburnea, while C. mastersii was sister to C. changningense and C. erythryeumn in section Iridorchis (). These information would provide genomic resources for the evolutionary status, taxonomy, and germplasm resources utilization in genus Cymbidium.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Chen SC. 1999. Cymbidium Sw. Vol. 18. In: Flora Reipublicae Popularis Sinicae. Beijing: Science Press; p. 191–227.
- Chen XQ. 2011. Manual of Chinese Cymbidiums and their cultivars. Beijing: China Forestry Publishing House.
- Cummings MP. 2004. PAUP* [phylogenetic analysis using parsimony (and other methods)]. Dictionary of bioinformatics and computational biology. John Wiley & Sons, Inc.
- Liu ZJ. 2006. Notes on some taxa of Cymbidium sect. Eburnea. Acta Phytotax Sin. 44(2):178.
- Liu ZJ, Chen SC, Cribb PJ. 2009. Orchidaceae. In: Wu ZY, Raven PH, Hong D, editors. Flora of China, vol. 25. Beijing: Science Press, Missouri Botanical Garden Press.
- Li Y, Li Z, Hu Q, Zhai J, Liu Z, Wu S. 2019. Complete plastid genome of Apostasia shenzhenica (Orchidaceae). Mitochondr DNA B. 4(1):1388–1389.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.