Abstract
The complete mitochondrial genome was sequenced from the marine monogonont rotifer Proales similis. The size of mitochondrial genome sequences was 16,819 bp and all the genes were unidirectional on the same strand. Of 12 protein-coding genes (PCGs), two genes (ND5 and ND2) had incomplete stop codons TG (for ND5) and T (for ND2). Furthermore, the start codon of all the genes was ATG, while the stop codons were TAA, TAG, and TGA. The base composition of 12 PCGs in P. similis mitogenome showed 20.8% for A, 47.0% for T, 17.4% for C, and 14.8% for G, respectively. The mitochondrial genome A + T base composition (67.8%) of 12 PCGs was higher than G + C (32.2%), while the complete mitochondrial genome A + T base composition (67.2%) was higher than G + C (32.8%).
To date, limited information is available on the complete mitochondrial genomes in the family Proalidae, while in the genus Brachionus, several research groups have published using the freshwater rotifers Brachionus calyciflorus (Choi et al. Citation2019) and Brachionus rubens (Choi et al. Citation2020a), while several marine rotifer Brachionus sp. mitogenomes were reported (Suga et al. Citation2008 for B. plicatilis; Hwang et al. Citation2014 for Brachionus koreanus, Kim et al. Citation2017 for Brachionus rotundiformis, and Choi et al. Citation2020b for Brachionus paranguensis).
The marine rotifer P. similis is one of the key aquaculture species as a dietary source for development of economically useful fish species such as seven-band grouper Epinephelus septemfasciatus (Wullur et al. Citation2009; Wullur et al. Citation2011; Hagiwara et al. Citation2014) with further development of this species for aquacultural physiology (e.g. salinity, microalgal density) (Rebolledo et al. Citation2018) and larviculure (Hagiwara and Marcial Citation2019). Among many rotifers, P. similis establishes yet another potential model for toxicity assessment of marine waters (Snell et al. Citation2019), implying that this species is useful for aquaculture and ecotoxicology. The sequence analysis of P. similis mitochondrial genome is also important to identify field-sampled and laboratory stocks. In this study, we identified complete mitochondrial genomes of the monogonont marine rotifer P. similis to better understand the phylogenetic placement of the marine rotifer Proales clade.
The adult P. similis were collected from Ishigaki island in Japan (24°25′N, 124°07′E) in July 2004 and maintained at the Laboratory of Professor Atsushi Hagiwara, Nagasaki University in Japan. The type was deposited in the ichthyological collection of the Faculty of Fisheries, Nagasaki University (FFNU) under the accession no. FFNU-Rot-0005. We sequenced 550 bp paired end (PE) library of P. similis from whole body genomic DNA using the Illumina HiSeq 2500 platform (GenomeAnalyzer, Illumina, San Diego, CA, USA). After the merge of two overlapping 6,495,659 PE reads into 228,955 single reads (average length 293 bp), de novo assembly was conducted by Newbler V2.9 (http://www.454.com) and 5228 contigs were obtained. Of them, five mitochondrial contigs were obtained. After a manual curation of five mitochondrial contigs with Consed (version 19.0) (http://www.phrap.org/consed/consed.html), one long contig was finally obtained to the mitochondrial DNA of P. similis. The complete mitochondrial genomes of P. similis were 16,819 bp in size (GenBank no. MN970216). Of 12 protein-coding genes (PCGs), two genes (ND5 and ND2) had incomplete stop codons TG (for ND5) and T (for ND2). Furthermore, the start codon of all the genes was ATG, while the stop codons were TAA, TAG, and TGA. The base composition of 12 PCGs in P. similis mitogenome showed 20.8% for A, 47.0% for T, 17.4% for C, and 14.8% for G, respectively. The mitochondrial genome A + T base composition (67.8%) of 12 PCGs was higher than G + C (32.2%), while the complete mitochondrial genome A + T base composition (67.2%) was higher than G + C (32.8%).
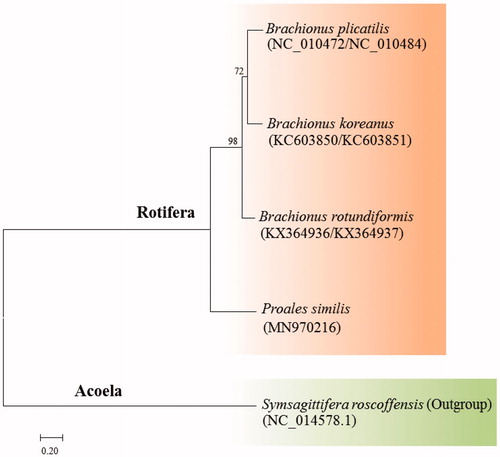
The phylogenetic placement of P. similis with 12 PCGs in the class Monogononta is shown in . The phylogenetic placement of marine rotifer P. similis is separated from the genus Brachionus spp. yet forms the cluster on more basal clade in the Rotifera (). In the family Proalidae, there was no report on complete mitochondrial genome; thus, comparative analysis of the gene order and contents of 12 PGCs with sister species in the same family was challenging, however, the differences in the gene order and contents of 12 PGCs were clear from Brachionus spp. in the family Brachionidae. This indicates that the complete mitochondrial genomes of rotifers are dynamic on gene structure and contents in the view of evolutionary event.
Figure 1. Phylogenetic analysis of the marine rotifer Proales similis mitochondrial DNA. We conducted a comparison of four monogonont rotifer species with 12 protein-coding genes (PCGs) with an outgroup. The amino acid sequences of 12 PCGs were aligned by ClustalW. Maximum likelihood (ML) analysis was performed by Raxml 8.2.8 (http://sco.h-its.org/exelixis/software.html) with GTR + γ+I nucleotide substitution model. The rapid bootstrap analysis was conducted with 1000 replications. The marine flatworm Symsagittifera roscoffensis served as an outgroup. Ln = −25855.214063.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Choi B-S, Lee YH, Hagiwara A, Lee J-S. 2019. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus calyciflorus (Rotifera, Brachionidae). Mitochondr DNA B. 4(2):3593–3595.
- Choi B-S, Lee YH, Lee J-S, Ogello EO, Kim H-J, Hagiwara A, Lee J-S. 2020a. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus rubens (Rotifera, Brachionidae). Mitochrondr DNA B. 5(1):5–6.
- Choi B-S, Kim D-H, Lee J-S, Kim H-J, Hagiwara A, Lee J-S. 2020b. Complete mitochondrial genome of the euryhaline monogonont rotifer Brachionus paranguensis (Rotifera, Brachionidae. Mitochondr DNA B. 5(1):502–503.
- Hagiwara A, Marcial HS. 2019. The use of non-Brachionus plicatilis species complex rotifer in larviculture. Hydrobiologia. 844(1):163–172.
- Hagiwara A, Wullur S, Marcial HS, Hirai N, Sakakura Y. 2014. Euryhaline rotifer Proales similis as initial live food for rearing fish with small mouth. Aquaculture. 432:470–474.
- Hwang D-S, Suga K, Sakakura Y, Hagiwara A, Park HG, Rhee J-S, Lee J-S. 2014. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera: Brachionidae). Mitochondr DNA. 25(1):29–30.
- Kim H-S, Hwang D-S, Kim H-J, Sakakura Y, Hagiwara A, Lee J-S. 2017. Complete mitochondrial genome of the monogonont rotifer Brachionus rotundiformis (Rotifera, Brachionidae. Mitochondr DNA B. 2(1):39–40.
- Rebolledo UA, Nandini S, Sarma SSS, Roman Reyes JC, Rodriguez Montes de Oca GA. 2018. Demographic and competition studies on Brachionus ibericus and Proales similis in relation to salinity and algal (Nannochloropsis oculata) density. Aquacult Int. 26(2):629–644.
- Snell TW, Johnston RK, Matthews AB, Park N, Berry S, Brashear J. 2019. Using Proales similis (Rotifera) for toxicity assessment in marine waters. Environ Toxicol. 34(5):634–644.
- Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. 2008. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 25(6):1129–1137.
- Wullur S, Sakakura Y, Hagiwara A. 2009. The minute monogonont rotifer Proales similis de Beauchamp: culture and feeding to small mouth marine fish larvae. Aquaculture. 293(1–2):62–67.
- Wullur S, Sakakura Y, Hagiwara A. 2011. Application of the minute monogonont rotifer Proales similis de Beauchamp in larval rearing of seven-band grouper Epinephelus septemfasciatus. Aquaculture. 315(3–4):355–360.
