Abstract
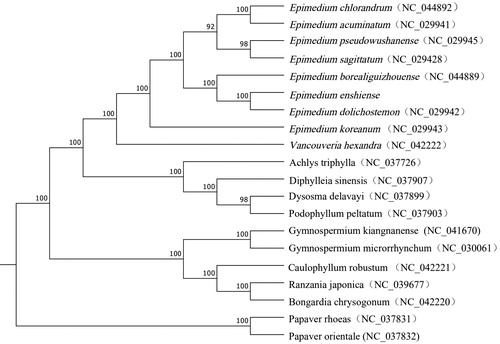
The genus of Epimedium belongs to Berberidaceae family, which is famous for their medicinal and ornamental value. In recent years, Epimedium has attracted increasing attention due to their medicinal and nutritive value. In this research, we reported the complete chloroplast (cp) genome of Epimedium enshiense. The complete chloroplast of this species is 157,076 bp in length, including a pair of invert repeat regions (IRS) (25,833 bp) that is divided by a large single copy area (LSC) (88,340 bp) and a small single copy area (SSC) (17,070 bp). The circular chloroplast genome of E. enshiense contains 112 unique genes, composing of 78 protein-coding genes, 30 tRNA, and four rRNA genes. Phylogenetic analysis indicates that E. enshiense has a closer relationship with E. dolichostmon.
Epimedium is a genus of herbaceous plants in the Berberidaceae family, which was used as traditional medicinal plants for more than 2000 years in China (Zhao et al. Citation2012). Epimedium enshiense B. L. Guo et Hsiao is a rare species with yellow spider-like flowers, which only inhabits in the Enshi County, Hubei Province, China (Guo and Xiao Citation1993). In this study, we reported the complete chloroplast sequence of E. enshiense, and analyzed the relationship between E. enshiense and other Epimedium species by phylogenetic analysis.
The sample of E. enshiense was collected from the Enshi County, Hubei province, China (E108°42, N28°26′). The voucher specimen (Guo0464) was deposited at the Herbarium of the Institute of Medicinal Plant (IMPLAD), Beijing, China. Total genomic DNA was extracted from the fresh leaves of E. enshiense with the modified CTAB method (Doyle and Doyle Citation1987). DNA library was sequenced, and 150 bp paired-end reads were generated on an Illumina Novaseq PE150 platform. The clean reads were assembled by using the program GetOrganelle v1.5 (Jin et al. Citation2018) with the reference chloroplast genome of E. acuminatum (GenBank accession number: KU522469). The chloroplast genome annotation was conducted through the online program CPGAVAS2 (Shi et al. Citation2019) and GeSeq (Tillich et al. Citation2017). The annotated chloroplast genomic sequence has been registered in GenBank with an accession number (MN937557).
The complete chloroplast genome of E. enshiense is 157,076 bp in length, and has a typical quadripartite construction, which contains two inverted repeat regions (IRa and IRb) of 25,833 bp that is insulated by a large single-copy (LSC, 88,340 bp) and a small single-copy (SSC, 17,070 bp). The total GC content of complete chloroplast genome, LSC, SSC, IR regions is 38.81%, 37.43%, 32.74% and 43.19%, respectively. The complete chloroplast genome of E. enshienseis contains 112 unique genes, including 78 protein-coding genes, 30 tRNA genes and four rRNA genes. Most of these genes are single-copy genes. However, four protein-coding genes (rps7, ndhB, ycf2, and rp123), seven tRNAs (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and four rRNAs (rrn16, rrn23, rrn4.5, and rrn5) are duplicated in the IR regions. One tRNA gene (trnQ-UUG) repeated in the LSC regions. In these genes, 15 genes (six tRNA genes and nine protein-coding genes) contain one intron, whereas three genes (ycf3, clpP, and rps12) contain double introns. The rps12 gene is trans-spliced, with the 5′ end located in the LSC region, and the 3′ end duplicated in the IR region.
To confirm the phylogenetic position of E. enshiense, the complete chloroplast genomes of 19 other plant species were downloaded from the NCBI GenBank database. The sequences were aligned using MAFFT v7 (Katoh et al. Citation2017), and then the maximum likelihood tree () was constructed using raxmlGUI1.5b (v8.2.10) (Silvestro and Michalak Citation2012). Phylogenetic analysis shows that E. enshiense is closely related to E. dolichostemon. The published E. enshiense chloroplast genome provides useful information for phylogenetic and evolutionary studies in Berberidaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Guo B, Xiao P. 1993. A new taxa of the genus Epimedium. J Syst Evol. 31:194–196.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 4:1–7.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhao Y, Song H, Fei J, Liang Y, Zhang B, Liu Q, Wang J, Hu P. 2012. The effects of Chinese yam-epimedium mixture on respiratory function and quality of life in patients with chronic obstructive pulmonary disease. J Tradit Chin Med. 32(2):203–207.