Abstract
The complete mitochondrial genome of Bartramia pomiformis (GenBank accession number MN972463) was 106,155 base pairs in length, containing 40 protein-coding genes, 3 ribosomal RNA (rRNA) and 24 transfer RNA (tRNA). The base composition was 30.5% A, 30.4% T, 20.2% G, 18.9% C and G + C 39.1%. The mitochondrial structure and gene order was similar to other Bryophytes. Phylogenetic tree based on the combined analysis of 33 protein coding genes was well congruent with traditional species relationship of the moss order Bartramiales.
The genus Bartramia consists of about 70 species in the world, of which 25 species are well-understood (Li et al. Citation2007). Bartramia pomiformis is one of the 13 species of common apple-mosses of the Bartramiaceae family known from South Korea (Park and Choi Citation2007). It is widely distributed in many habitats globally, but most often found in temperate areas (Li et al. Citation2007) and on rocks covered by thin soil, humus, rotten trees or tree trunks in Korea (Choe Citation1980).
Bartramia pomiformis specimens for complete mitogenome sequencing were collected from a 3 × 3 cm patch of a population growing under natural conditions of Yeongyang-gun County, Gyeongsangbuk-do Province (36° 38′ 19.04 N; 129° 9′ 24.82 E) on 21 October 2019. The specimen was deposited into the JNU Herbarium in Chonju, Korea with the accession number YYJ 20191021-1. The genomic DNA was extracted from fresh plants using the DNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) and a sequencing library was prepared using the TruSeq Nano DNA Library Prep Kit (Illumina, San Diego, CA, USA). Genome sequencing was carried out by the Illumina HiSeq 2 × 150 bp Sequencing System (Illumina). Trimming and filtering of raw reads was performed using the FastQC v0.11.5 (Andrews Citation2010) and the Trimmomatics v0.36 tools (Bolger et al. Citation2014). Filtered reads were then assembled into contigs in the Geneious 8.1.9 (Biomatters Ltd., Auckland, New Zealand) and NOVOplasty 2.4 software (Dierckxsens et al. Citation2016). Genome annotation was performed using the Geneious 8.1.9 software based on B. pomiformis (NC_024519).
The mitogenome of B. pomiformis is 106,155 bp long and consists of 40 protein-coding genes, 3 ribosomal RNAs (rRNAs) and 24 transfer RNAs (tRNAs). The base composition is: 30.5% A, 30.4% T, 18.9% C, 20.2% G, 60.9% AT, and 39.1% GC. The mitochondrial structure and gene order are similar to that of other Bryophytes.
To identify the position of B. pomiformis within the Bryophytes, a phylogenetic tree was constructed by aligning the amino acid sequences of 33 mitochondrial genes of B. pomiformis with those from 21 Bryophyta and 3 Marchantiophyta species available in the public database. All downstream analyses were performed using the program MEGA 7 based on maximum-likelihood (ML) analysis with the JTT matrix-based model () (Tamura et al. Citation2013). Supports for internal branches were tested using the bootstrap analyses of 1000 replications in ML analyses. Analysis of B. pomiformis dataset for the order Bartramiales provides a convincing support for many traditionally-recognized genera and identifies a higher level of phylogenetic structure () (Cox et al. Citation2000; Magombo Citation2003; Liu et al. Citation2014).
Figure 1. Phylogenetic position of B. pomiformis determined by Maximum likelihood methods based on combined analysis with amino acids sequences of 33 mitochondrial genes common in all taxa. The bootstrap values (1000) are presented near the corresponding branch. Branches that were supported by above 50% bootstrap values are indicated by bold lines. Sequences from Marchantiopsida were used as outgroup. GenBank accession numbers of mitogenomes used are Anomodon attenuatus (NC_021931), Anomodon rugelii (NC_016121), Atrichum angustatum (NC_024520), Bartramia pomiformis (NC_024519), Buxbaumia aphylla (NC_024518), Climacium americanum (NC_024515), Climacium dendroides (MN942036), Codriophorus laevigatus (NC_025931), Funaria hygrometrica (NC_024523), Hypnum imponens (NC_024516), Marchantia polymorpha (NC_001660), Mielichhoferia elongate (NC_036945), Orthotrichum speciosum (NC_026121), Orthotrichum stellatum (NC_024522), Physcomitrella patens (NC_007945), Pleurozia purpurea (NC_013444), Ptychomnion cygnisetum (NC_024514), Racomitrium ericoides (NC_026540), Sphagnum palustre (NC_024521), Syntrichia filaris (KP984758), Tetraphis pellucida (NC_024290), Tetraplodon fuegianus (NC_028191), Treubia lacunosa (NC_016122), and Ulota hutchinsiae (NC_024517).
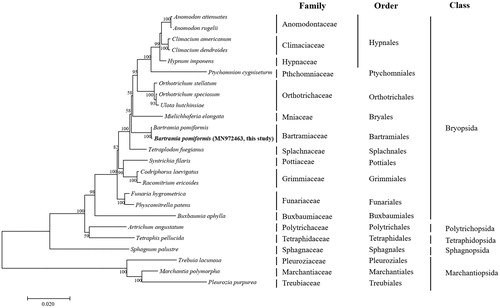
Until recently, over 60 mitogenomes from Bryophyta have been reported (Kang et al. Citation2019), but this is the first report of a moss flora from Korea belonging to order Bartramiales. Although the mitogenome of B. pomiformis sampled in America is already reported (Liu et al. Citation2014), this study will enable a better understanding of the molecular phylogeographical studies and the evolution of mosses displaying disjunct distributions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data; [accessed 2020 Feb 13]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Choe D.M. 1980. Illustrated Flora and Fauna of Korea. Vol. 24. Musci-Hepaticae. Seoul: Ministry of Education. p. 790.
- Cox CJ, Goffinet B, Newton AE, Shaw AJ, Hedderson TA. 2000. Phylogenetic relationships among the diplolepideous-alternate mosses (Bryidae) inferred from nuclear and chloroplast DNA sequences. The Bryologist. 103(2):224–241.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kang P, Cho S. M, Lee J, Yim J. H, Lee J. H, Lee H. 2019. The complete mitogenome of the Arctic moss Aulacomnium turgidum (Wahlenb.) Schwaegr. Mitochondrial DNA Part B. 4(2):3446–3447.
- Li X. J, Crosby M. R, Si H. 2007. Moss Flora of China. Vol. 4. Beijing, New York and St. Louis: Science Press and Missouri Botanical Garden Press. p. 211.
- Liu Y, Rafael M, Goffinet B. 2014. 350 million years of mitochondrial genome stasis in mosses, and early land plant lineage. Mol. Biol. Evol. 31(10):2586–2591.
- Magombo ZL. 2003. The phylogeny of basal peristomate mosses: evidence from cpDNA, and implications for peristome evolution. Syst. Bot. 28(1):24–38.
- Park K.-W, Choi K. 2007. New list of bryophytes of Korea. Pocheon (in Korean): Korea National Arboretum. p. 150.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30(12):2725–2729.
