Abstract
Indian rose-ringed parakeet (Psittacula krameri) is one of the most recognizable illegally trafficked wild birds. With feral populations in Europe and Australia, its phylogeny and taxonomy are of great interest amongst biologists. The information regarding the genetic composition and make up of Psittacula species from various geographic regions is scarce and incomplete. This study aimed to generate the first partial mitogenome sequence of the species from India. We generated 2611 base pair long mitochondrial sequence of P. krameri. The overall base composition was calculated at AT (52.59%) and GC (47.41%). The complete NAD1 protein-coding gene was annotated along with partial NAD2 protein-coding gene. The sequence encoded four tRNA genes (Isoleucine, Leucine, Glutamine, and Methionine) as well as partial sequence of 16 s rRNA. Psittacula krameri demonstrated closer phylogenetic relation with P. eupatria than P. derbiana. We also reported that Psittacula species have closest phylogenetic relationship with E. roratus than any other parrots.
The genus Psittacula of Psittacidae family is a group of medium-sized (∼40 cm) birds found naturally in a variety of habitats across Asia and Africa comprising 10–15 extant species (Groombridge et al. Citation2004). Owing to the popularity of the species as a cage bird, rose-ringed parakeet (P. krameri) has also established feral populations across Europe and Australia (BirdLife International Citation2016). Though fairly common, the information regarding the genetic composition and make up of Psittacula species is scarce and incomplete (Liu et al. Citation2019). Hence, more information on mitogenome of Psittacula species from various geographical locations is very much warranted, and through this study we attempted to redress the same. To date, no report is available about complete/partial mitogenome of Indian rose-ringed parakeet exclusively from the Indian subcontinent though a complete mitochondrial genome of P. krameri from Australia is published recently (Sarker et al. Citation2019). We attempted to sequence the complete mitochondrial genome of the Indian rose-ringed parakeet from India using multiple primers and were successful in retrieving information about partial mitochondrial genome of the above-mentioned species.
Shed contour feathers of P. krameri were collected from Sri Chamarajendra Zoological Gardens, Mysuru, Karnataka (12°18′06″N, 76°39′50″E) with due permission and used for the study. The DNA sample is deposited in the Avian Genome Resource Bank of National Avian Forensic Laboratory at Sálim Ali Center for Ornithology and Natural History under the accession number NAFL/0150.2/DNA/140519. DNA extraction was done from the feather calamus by means of Phenol-Chloroform-Isoamyl Alcohol method with minor modifications (Sambrook et al. Citation1989). A set of 18 primers were designed and used, of which, only 5 resulted in amplification from mitochondrial DNA (Supplementary Tables 1 and 2). The PCR amplified amplicons were gel purified and cycle sequenced with Big Dye Terminator ver. 3.1 (Applied Biosystems, Foster City, CA) using Applied Biosystems Genetic Analyzer 3500 (Applied Biosystems, Foster City, CA). The sequences generated by ABI 3500 were checked using Sequence Analysis Software Version 6.0 (Applied Biosystems, Foster City, CA) for quality control. Sequence alignment and gap filling were carried out using Unipro UGENE (Okonechnikov et al. Citation2012). Annotation was carried out by comparing the gene map created with Mitos Web-server and aligning it manually with the already published P. krameri (GenBank accession: MN065674) mitogenome, and the results of tRNA were verified in tRNAScanSe web portal.
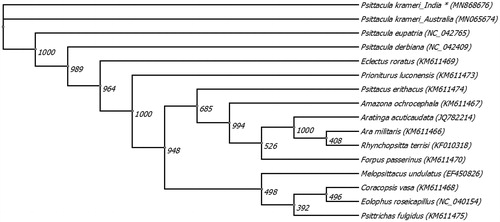
This study reported a 2611 base pair long sequence of P. krameri mitogenome with GenBank accession number MN868676. The overall base composition of the sequence was dominated by higher AT (52.59%) than GC (47.41%) content. The contents of A, T, G, and C were 31.1%, 33.6%, 21.5%, and 13.9%, respectively. The sequence encoded NADH dehydrogenase subunit I (NAD1) complete gene (981 bases), partial sequence of 16S ribosomal RNA (rRNAL, 681 bases), and NADH dehydrogenase subunit I (NAD2) gene (657 bases) along with four complete tRNA genes of Leucine (TTA), Isoleucine (ATC), Glutamine (CAA), and Methionine (ATG). Phylogenetic analysis was performed on sequenced P. krameri mitochondrial genome regions (MN868676), the published P. krameri mitogenome (MN065674) and 14 other parrot species (). The region of maximum alignment was extracted and analyzed by means of maximum likelihood method using 1000 bootstrap replicates in Unipro UGENE Estimates of evolutionary divergence between the sequences were conducted using the Kimura 2-parameter model (Kimura Citation1980) employing default settings in MEGA X (Kumar et al. Citation2018). The genetic divergence between the sequenced P. krameri (MN868676) and published P. krameri (MN065674) was calculated at 0.3%. P. krameri was comparatively closer to P. eupatria than P. derbiana and the three Psitaculla species are closest to E. roratus corroborating previous studies (). We concluded that P. krameri being one of the most widely introduced and trafficked species of birds, this data set will be a useful tool for studying various evolutionary questions of Psittacula species and/or designing conservation and management strategies for the species including its application in illegal avian trade.
Supplemental Material
Download MS Word (25.1 KB)Acknowledgements
We are thankful to the Forest Department of Karnataka and Mr. Ajit Kulkarni IFS (Executive Director, Sri Chamarajendra Zoological Gardens, Mysuru, Karnataka), for providing permission (No. MZA/62/2019-20) to collect shed feathers for this study.
Disclosure statement
No potential conflict of interests was reported by the author(s).
Additional information
Funding
References
- BirdLife International. 2016. Psittacula krameri. IUCN Red List of Threatened Species. Version 2016.3. International Union for Conservation of Nature. [accessed 18 June 2018].
- Groombridge J. J, Jones C. G, Nichols R. A, Carlton M, Bruford M. W. 2004. Molecular phylogeny and morphological change in the Psittacula parakeets. Mol Phylogenet Evol. 31(1):96–108.
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. (2)16:111–120.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (6)35:1547–1549.
- Liu H, Ding Y, Chen R, Tong Q, Chen P. 2019. Complete mitogenomes of two Psittacula species, P. derbiana and P. eupatria. Mitochondrial DNA B. 4(1):692–693.
- Okonechnikov K, Golosova O, Fursov M, Ugene T. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 28(8):1166–1167.
- Sambrook J, Fritsch E. F, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- Sarker S, Sutherland M, Talukder S, Das S, Forwood JK, Helbig K, Raidal SR. 2019. The first complete mitogenome of Indian ringneck (Psittacula krameri) demonstrates close phylogenetic relationship with Eclectus parrot. Mitochondrial DNA B. 4(2):3579–3581.