Abstract
Lonicera maximowiczii (Caprifoliaceae) is a deciduous shrub with great value for its decorative leaves and colorful flowers, which has massively cultivated in parks and gardens for ornamental purposes. Here, the complete chloroplast genome of the L. maximowiczii been constructed from the Illumina sequencing data. The circular cp genome is 155,584 bp in size and comprises a pair of inverted repeat (IR) regions of 23,791 bp each, a large single-copy (LSC) region of 88,056 bp, and a small single-copy (SSC) region of 19,946 bp. The total GC content is 38.4%, whereas the corresponding values of the LSC, SSC, and IR region are 36.9%, 33.3%, and 43.4%, respectively. The chloroplast genome contains 129 genes, including 81 protein-coding genes, eight ribosomal RNA genes, and 39 transfer RNA genes. The Maximum-Likelihood Phylogenetic analysis showed a strong sister relationship with Lonicera macranthoides. Our findings can be subsequently used for population, phylogenetic, and chloroplast genetic engineering studies in Lonicera.
Lonicera maximowiczii belongs to Caprifoliaceae family is a deciduous shrub that grows up to 2 m in height in forests or forest margins at an altitude of 800–1800 m. It is distributed in northeastern China (i.e. the Xiaoxinganling Mountains in Heilongjiang, the Changbai Mountains of Jilin, and to the eastern end of the Liaodong Peninsula), northern Korea and the Russian Far East. Flowers and fruits are ornamental, and the strong germination ability of L. maximowiczii can be used as landscape plants in flower borders and beds, also as honey plants. In addition, it is often used for physical fitness due to its massive biological compounds and their pharmaceutical properties (Rupasinghe et al. Citation2018; Molina et al. Citation2019). Despite its horticultural and economic importance, few genetic and genomic research have been done on this plant. Therefore, we reported the complete chloroplast genome (cp) of L. maximowiczii based on Illumina sequencing data, which would be helpful for its evolution and genetics research.
Fresh leaves of L. maximowiczii were collected from Baishi Mountain in Jilin city, Jinlin province of China (126°46′43″E, 43°24′47″N) for total genomic DNA extraction. The voucher specimen was preserved at the Jilin Engineering Vocational College (accession number BBRD18058). The whole-genome sequencing was conducted with 150 bp pair-end reads on the Illumina Hiseq Platform and totally 1.6 Gb were obtained and then de novo assembled using CLC Genomics Workbench v7.5 (CLC Bio, Aarhus, Denmark) (Kim et al. Citation2015). A subset of 59.81 M trimmed reads were used for constructing the chloroplast genome by MITObim v1.8 (Hahn et al. Citation2013), with that of its congener Lonicera japonica (GenBank: NC_026839.1) as the initial reference genome. The chloroplast genome was annotated in GENEIOUS R9 (Biomatters Ltd., Auckland, New Zealand). Finally, the validated complete chloroplast genome sequence was submitted to GenBank with accession number MN986996.
The complete chloroplast genome of L. maximowiczii is 155,584 bp in size with high coverage (mean 372.2×), containing a pair of inverted repeat (IR) regions of 23,791 bp each, separated by a large single-copy (LSC) region of 88,056 bp and a small single-copy (SSC) region of 19,946 bp. The total GC content is 38.4%, whereas the corresponding values of the LSC, SSC, and IR region are 36.9%, 33.3%, and 43.4%, respectively. This chloroplast genome harbours 129 functional genes, including 81 protein-coding genes, 39 tRNA genes, and eight rRNA genes. Protein-coding genes, rps7, ndhB, and ycf2, were duplicated genes which located in IR regions. Besides, 18 genes contain one intron, whereas chlp, rps12, and ycf3 harbour two introns. Summarily, the L. maximowiczii cp genome is structurally similar to previously published genomes (He et al. Citation2017; Hu et al. Citation2018).
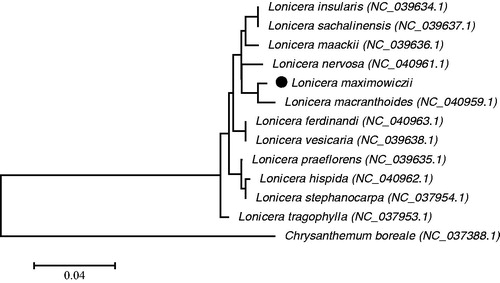
The phylogenetic tree was generated based on the common coding protein genes of L. maximowiczii and the other 10 species (). The alignment was conducted using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was built using the maximum-likelihood (ML) method. The results showed that L. maximowiczii was closely related to Lonicera macranthoides. Our findings can be subsequently used for population, phylogenetic, and chloroplast genetic engineering studies in Lonicera.
Figure 1. The phylogenetic tree was constructed using chloroplast genome sequences of 12 species in Lonicera and setting Chrysanthemum boreale as outgroup based on the Maximum-Likelihood analysis using 500 bootstrap replicates. Chloroplast genome sequences used for this tree are Chrysanthemum boreale, NC_037388.1; Lonicera ferdinandi, NC_040963.1; Lonicera hispida, NC_040962.1; Lonicera insularis, NC_039634.1; Lonicera maackii, NC_039636.1; Lonicera macranthoides, NC_040959.1; Lonicera nervosa, NC_040961.1; Lonicera praeflorens, NC_039635.1; Lonicera sachalinensis, NC_039637.1; Lonicera stephanocarpa, NC_037954.1; Lonicera tragophylla, NC_037953.1; Lonicera vesicaria, NC_039638.1.
Disclosure statement
The authors report no conflicts of interest, and alone are responsible for the content and writing of the paper.
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- He L, Qian J, Li X, Sun Z, Xu X, Chen S. 2017. Complete chloroplast genome of medicinal plant Lonicera japonica: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules. 22(2):249.
- Hu H, Liu JG, An JX, Wang M, Wang Q. 2018. Characterization of the complete chloroplast genome of Lonicera macranthoides. Mitochondrial DNA B. 3(2):1000–1001.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast ad ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5(1):15655.
- Molina AK, Vega EN, Pereira C, Maria ID, Sandrina AH, Paula R, Isabel PF, Maria FB, Marina K, Marina S, et al. 2019. Promising antioxidant and antimicrobial food colourants from Lonicera caerulea L. var. kamtschatica. Antioxidants. 8(9):394.
- Rupasinghe HPV, Arumuggam N, Amararathna M, De Silva ABKH. 2018. The potential health benefits of haskap (Lonicera caerulea L.): role of cyanidin-3-O-glucoside. J Funct Foods. 44:24–39.
