Abstract
The Hestina assimilis belongs to Nymphalidae in Lepidoptera. We described the complete mitogenome of H. assimilis, which is typical circular duplex molecules and 15,262 bp in length, containing the standard metazoan set of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and an A + T-rich region with macro-repeat sequences. All the inferred tRNA secondary structures show the common cloverleaf pattern, with the exception of trnS1(AGN) which lacks the DHU arm. The H. assimilis mitochondrial genome has the same gene order as other lepidopterans.
The butterfly Hestina assimilis (Lepidoptera: Nymphalidae) is a forestry pest. Its large larval occurrence mainly harms Ulmaceae plants such as Celtis sinensis. Such damage leads to consider quantitative and qualitative economic losses (Kato Citation2007). Here, we firstly sequenced the complete mitochondrial genome of H.assimilis based on the samples collected from Shanxi province in China (37°83′33″N, 112°66′61″E), which would facilitate our understanding of the mitochondrial genome, phylogeography and the phylogenic relationship of the Nymphalidae family.
The adult stage of H.assimilis was washing with 70% ethanol first, and then those were stored in 95% ethanol. The mitochondrial DNA was extracted using De Novo sequencing library. The voucher specimen’s genome DNA is deposited in the Shanxi Insect Herbarium, Shanxi Academy of Agricultural Sciences, and their number is 2018TY501-510. The complete sequence of H.assimilis mitochondrial genome (GenBank accession no. MN182752) is 15,262 bp in length, consisting of 13 protein-coding genes, two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a control region. Twenty-three genes (trnI, trnM, nad2, trnW, cox1, trnL2, cox2, trnK, trnD, atp8, atp6, cox3, trnG, nad3, trnA, trnR, trnN, trnS1, trnE, trnT, nad6, cytB, and trnS2) were found to be encoded by the majority strand (J-strand) and the other 14 genes (trnQ, trnC, trnY, trnF, nad5, trnH, nad4, nad4L, trnP, nad1, trnL1, rrnL, trnV, and rrnS) by minority strand (N-strand), those similar to other butterflies (Wu et al. Citation2016). Overall nucleotide compositions are 38.76% of A, 11.92% of C, 41.67% of T, and 7.62% of G, with an AT content of 80.43%. Most protein-coding genes begin with ATN, such as cox2, atp6, cox3, cytb, nad1, nad4, nad4L, and nad6 genes employing ATG, whereas cox1 starts with CGA, the rest use ATT as a start codon. And, the exception of nad4, nad5, and cox2, which terminates with a single T—, nad4L and nad3 are terminated with TAG, others with TAA as the stop codon. The rrnL is located between trnL1 and trnV, whereas rrnS is between trnV and control region. The secondary structure of tRNAs exhibited typical clover-leaf structure is same as other insects. The control region is located between rrnS and trnM. The gene order model was same as other reported insects (Garey and Wolstenholme Citation1989).
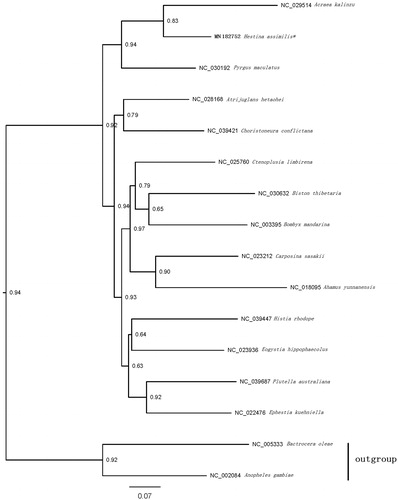
The sequences were aligned with MAFFT v7.2 software (Katoh and Standley Citation2013); the evolutionary analyses were conducted with RAxML v8.2.10 (Stamatakis Citation2014) on the CIPRES Science Gateway (Miller et al. Citation2010). The phylogenetic relationships of the H. assimilis mitogenome assembly references and 15 other Lepidoptera taxa from GenBank, were reconstructed based upon the concatenated nucleotide sequences of the 13 PCGs (). The phylogenetic tree shows that H. assimilis is part of the Nymphalidae, all members of which are clustered into a clade.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of H. assimilis was deposited in GenBank under the accession number MN182752.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Garey JR, Wolstenholme DR. 1989. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 28(5):374–387.
- Kato. 2007. An attempto to clarify environmental stimuli responsible for the production of white morphs in a nymphalid butterfly Hestina assimilis. Butterflies. 45:12–19.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans, Louisiana: Institute of Electrical and Electronics Engineers (IEEE); p. 1–8.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu YP, Zhao JL, Su TJ, Luo AR, Zhu CD. 2016. The complete mitochondrial genome of Choristoneura longicellana (Lepidoptera: Tortricidae) and phylogenetic analysis of Lepidoptera. Gene. 591(1):161–176.