Abstract
Clam shrimps (Spinicaudata) are a widespread and diverse crustacean group that frequent temporary aquatic habitats, but few complete mitochondrial genomes have been published for this group. Here, we report the mitogenome of an undescribed Gondwanalimnadia species from Botswana. Raw sequences were assembled into a single circular genome with a total length of 15,663 bp. Thirteen protein-coding genes, 22 tRNAs, and 2 rRNAs were identified using the MITOS pipeline. The mitogenome’s GC content is 33.52%. Phylogenetic analysis using protein-coding genes confirmed that Gondwanalimnadia sp. is closely related to another member of the Limnadiidae, Limnadia lenticularis.
Spinicaudatans are an order of freshwater crustaceans commonly known as spiny clam shrimps. These large branchiopod crustaceans occur exclusively in seasonally astatic aquatic habitats (Brendonck et al. Citation2008). Similar to other branchiopods, clam shrimp rapidly attain sexual maturity and within short hydroperiod windows, produce dormant eggs that can withstand dry periods (Brendonck et al. Citation2008; Rogers Citation2009). They are thought to be omnivores with diets consisting of detritus, plankton and algae (Hethke et al. Citation2019). Gondwanalimnadia spp. belong to the Limnadiidae (Weeks et al. Citation2012; Rogers et al. Citation2016), a family in the order Spinicaudata with a global distribution, except for Antarctica (Bellec and Rabet Citation2016). As with many spinicaudatan families, little is known about their genomic make-up, and their correct placement within the branchiopod phylogeny is not fully resolved (Bellec and Rabet Citation2016). Here, we describe the complete mitogenome of an undescribed Gondwanalimnadia species obtained from Botswana, and investigate its phylogenetic placement among several other brachiopods.
Specimens of the undescribed Gondwanalimnadia species were collected from a temporary pond on the outskirts of Palapye, Central district, Botswana (GPS 27.16616 E, 22.54507 S) and preserved in 80% ethanol. Voucher specimens from the locality were deposited at the Kansas Biological Survey (DCR-1138). Genomic DNA of high molecular weight was extracted using the CTAB method (Doyle and Doyle Citation1987). An indexed DNA library was constructed using NEBNext DNA Library Preparation Kit (Massachusetts, USA), and 1 µg of genomic DNA as template. The DNA library was sequenced on the Illumina Hi-Seq platform (California, USA) using 2 × 150 chemistry with an average insert size of 350 bp.
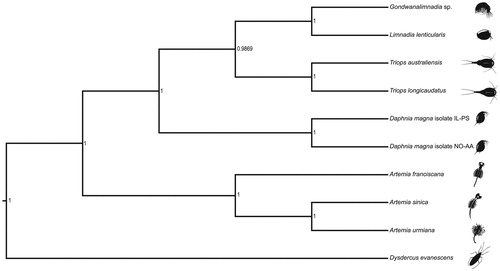
The Illumina sequencing run yielded a total of 23,096,464 raw sequences. The complete mitogenome was assembled using NOVOPlasty v3.5 (Dierckxsens et al. Citation2016) and annotated using MITOS Web Server (Bernt et al. Citation2013). The assembly resulted in a single circular genome with a total length of 15,663 bp. The MITOS pipeline identified 13 protein-coding genes, 22 tRNAs, and 2 rRNAs, typical of all crustaceans. The GC content of the assembly was estimated at 33.52%. Several instances of non-canonical start codon and incomplete stop codons were observed, as for other arthropods (Monsanto et al. Citation2019; Jagatap et al. Citation2019). Protein coding sequences from the studied species, eight closely related species with complete mitogenomes in the NCBI database, and an outgroup (Dysdercus evanescens) were aligned in MAFFT v7.429 (Katoh et al. Citation2009). A Bayesian phylogenetic tree was reconstructed using package BEAST2 (Bouckaert et al. Citation2014). The program’s default settings were used, except that the substitution model was set to HKY (Hasegawa et al. Citation1985) with four gamma categories. BEAST2 was run for 50,000,000 iterations with 30% burn-in. Markov chain convergence and Effective Sample Size (ESS) were visually inspected in Tracer v1.7 (Rambaut et al. Citation2018). The phylogenetic tree was visualized in FigTree v1.4 (Rambaut and Drummond Citation2012), placing the uncharacterized Gondwanalimnadia sp., along with another limnadiid (Limnadia lenticularis), in a monophyletic clade with cladoceran and notostracan representatives ().
Figure 1. A Bayesian phylogenetic tree constructed in BEAST2 using mitogenome sequences of an undescribed Gondwanalimnadia species (NCBI accession number MN625703) and nine related species. Daphnia magna isolate IL-PS: MH683649.1, Daphnia magna isolate NO-AA: MH683655.1, Limnadia lenticularis: NC_039394.1, Triops australiensis: LK391946.1, Triops longicaudatus: AY639934.1, Dysdercus evanescens: NC_042437.1, Artemia urmiana: NC_021382.1, Artemia sinica: NC_042147.1, Artemia franciscana: X69067.1. The numbers on the tree indicate the posterior probability estimated for each node.
Disclosure statement
The authors declare that there is no existing competition and/or financial interest . Therefore, opinions, findings, conclusions or recommendations expressed in this material are owned by the authors.
Additional information
Funding
References
- Bellec L, Rabet N. 2016. Dating of the Limnadiidae family suggests an American origin of Eulimnadia. Hydrobiologia. 773(1):149–161.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 10(4):e1003537.
- Brendonck L, Rogers DC, Olesen J, Weeks S, Hoeh WR. 2008. Global diversity of large branchiopods (Crustacea: Branchiopoda) in freshwater. Hydrobiologia. 595(1):167–176.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18–e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22(2):160–174.
- Hethke M, Fürsich FT, Jiang B, Wang B, Chellouche P, Weeks SC. 2019. Ecological stasis in Spinicaudata (Crustacea, Branchiopoda)? Early Cretaceous clam shrimp of the Yixian formation of north-east China occupied a broader realized ecological niche than extant members of the group. Palaeontology. 62(3):483–513.
- Jagatap H, Monsanto DM, Jansen van Vuuren B, Janion-Scheepers C, Sekar S, Teske PR, Emami-Khoyi A. 2019. The complete mitogenome of the springtail Tullbergia bisetosa: a subterranean springtail from the sub-Antarctic region. Mitochondrial DNA B. 4(1):1594–1596.
- Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. In: Posada D, editor. Bioinformatics for DNA sequence analysis. Methods in molecular biology (methods and protocols). Clifton, NJ: Humana Press; vol 537. p. 39–64.
- Monsanto DM, Jansen van Vuuren B, Jagatap H, Jooste CM, Janion-Scheepers C, Teske PR, Emami-Khoyi A. 2019. The complete mitogenome of the springtail Cryptopygus antarcticus travei provides evidence for speciation in the Sub-Antarctic region. Mitochondrial DNA B. 4(1):1195–1197.
- Rambaut A, Drummond AJ. 2012. FigTree: tree figure drawing tool, v1.4.2. Edinburgh: Institute of Evolutionary Biology, University of Edinburgh. [accessed 2020 Feb 4]. http://tree.bio.ed.ac.uk/software/figtree/
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biol. 67(5):901–904.
- Rogers DC. 2009. Branchiopoda (Anostraca, Notostraca, Laevicaudata, Spinicaudata, Cyclestherida). In: Likens GF, editor. Encyclopedia of inland waters. Amsterdam: Elsevier; Vol. 2, p. 242–249.
- Rogers DC, Rabet N, Weeks SC. 2016. Gondwanalimnadia (Branchiopoda: Spinicaudata), replacement name for Afrolimnadia Rogers, Rabet and Weeks, 2012 (Limnadiidae), junior homonym of Afrolimnadia Tasch, 1987 (Lioestheriidae). J Crustacean Biol. 36(1):105–105.
- Weeks SC, Rogers DC, Rabet N. 2012. Revision of the extant genera of Limnadiidae (Branchiopoda: Spinicaudata). J. Crustacean Biol. 32:827–842.
