Abstract
This paper reports the mitochondrial genome of Calidris pugnax, which is a closed circular molecule of 16,902 bp. The overall base composition of this species is 25.2% T, 30.6% C, 32.0% A, and 12.2% G, with an A + T content of 55.2%. The structure of the mitogenome is a typical vertebrate mitochondrial gene arrangement. Phylogenetic analysis of complete mitochondrial genome concatenated sequences from 13 species from 6 genera was conducted using the maximum-likelihood (ML) model. The topology demonstrated that the mitogenome of this species was genetically closest to that of Calidris tenuirostris. Calidris pugnax mitogenome can contribute to our understanding of the phylogeny and evolution of this species.
Ruff (Calidris pugnax), which belongs to the genus Calidris within the family Scolopacidae (Aves, Charadriiformes), mainly breeds in Northern Eurasia and migrates to Africa, southeast Asia, and Australia during winter. Ruff is a single species with no subspecies, and was placed in monotypic genus Philomachus before, but a recent study indicates that the latter is embedded within Calidris (Gibson and Baker Citation2012). Ruff is special among Scolopacidae bird species, and the morphology of males are diverse during the breeding period. In this study, we determined the complete mitochondrial genome (mt genome) of C. pugnax for the first time. The samples were collected at the Bird Circle Station of the Guiyang Longdongbao International Airport (26°32.983′N, 106°48.585′E), Guizhou province, China in December 2019. After sampling, the specimens (NJFU-2020-LSY) were stored in the Animal Specimens Museum of Nanjing Forestry University. Genome DNA was isolated from paw tissue using a kit and 12 primers were designed for fragment amplification.
The complete mitochondrial genome of C. pugnax is a closed circular molecule composed of 16,902 bp (GenBank accession no. MN956840), containing the typical set of 13 protein-coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes (rrnS and rrnL), and one control region (D-loop). The nucleotide composition is 25.2% T, 30.6% C, 32.0% A, and 12.2% G, with an A + T content of 55.2%. The structure of the mitogenome is a typical vertebrate mitochondrial gene arrangement (Noack et al. Citation1996; Liu, Yang et al. Citation2019; Sun et al. Citation2019).
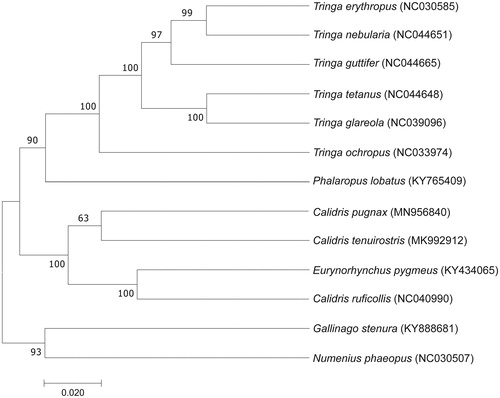
In this study, phylogenetic analysis of the C. pugnax and 13 other Scolopacidae bird species was carried out based on the concatenated nucleotide sequences of the complete mitochondrial genome using the maximum-likelihood (ML) model in MEGA 7.0 (Kumar et al. Citation2016) with 1000 bootstrap replicates. In this phylogenetic tree, C. pugnax is clustered with C. tenuirostri, which indicates that they have a much closer relationship (), The phylogenetic tree reveals that the data of our new determined mitogenome explain some related evolution issues. It appears that C. pugnax is more closely related to the birds of the genus Calidris, the phylogenetic analysis result was consistent with the previous research (Gibson and Baker Citation2012). Calidris pugnax mitogenome can contribute to our understanding of the phylogeny and evolution of this species. (Boore Citation1999; Liu, Xu et al. Citation2019; Sun et al. Citation2020).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Gibson R, Baker AJ. 2012. Multiple gene sequences resolve phylogenetic relationships in the shorebird suborder Scolopaci (Aves: Charadriiformes). Mol Phyl Evol. 64(1):66–72.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu H, Xu N, Yang J, Zhang Q, Zuo K, Lv F. 2019. The complete mitogenome of Hong Kong paradise fish (Macropodus hongkongensis), an endemic freshwater fish in South China. Mitochondrial DNA Part B. 4(2):2849–2850.
- Liu H, Yang Y, Xu N, Chen R, Fu Z, Huang S. 2019. Characterization and comparison of complete mitogenomes of three parrot species (Psittaciformes: Psittacidae). Mitochondrial DNA Part B. 4(1):1851–1852.
- Noack K, Zardoya R, Meyer A. 1996. The complete mitochondrial DNA sequence of the bichir (Polypterus ornatipinnis), a basal ray-finned fish: ancient establishment of the consensus vertebrate gene order. Genetics. 144(3):1165–1180.
- Sun CH, Liu B, Lu CH. 2019. Complete mitochondrial genome of the Siberian thrush, Geokichla sibirica sibirica (Aves, Turdidae). Mitochondrial DNA Part B. 4(1):1150–1151.
- Sun CH, Liu HY, Lu CH. 2020. Five new mitogenomes of Phylloscopus (Passeriformes, Phylloscopidae): sequence, structure, and phylogenetic analyses. Int J Biol Macromol. 146:638–647.