Abstract
The whole chloroplast (cp) genome sequence of Viburnum schensianum has been characterized from Illumina pair-end sequencing. The complete cp genome was 158,408 bp in length, containing a large single copy region (LSC) of 86,998 bp and a small single copy region (SSC) of 18,386 bp, which were separated by a pair of inverted repeat (IR) regions of 26,512 bp. The genome contained 130 genes, including 84 protein-coding genes, 37 tRNA genes, eight ribosomal RNA genes (four rRNA species), and a pseudogene. Most genes occur as a single copy, whereas 16 gene species are duplicated. Phylogenetic analysis revealed that V. schensianum is closely related to the species of V. utile.
Genus Viburnum belonging to family Adoxaceae consists of about 200 species of shrubs and small trees (Lobstein et al. Citation1999); most Viburnum species have been used in medicines because flavonoids, bioflavonoids, and coumarins are reported in different Viburnum species (Cometa et al. Citation1998). Viburnum schensianum is an important species in Viburnum which widely distributed in central and southern of China at an altitude between 700 and 2200 m(Wang Citation2006). In the past few years, some researches focus on genetic relationships of Viburnum species based on chloroplast genome or gene fragments (Donoghue et al. Citation2004; Donoghue and Winkworth Citation2005; Bibi et al. Citation2010; Clement et al. Citation2014). However, there have been no genomic studies on Viburnum schensianum. Herein, we assembled the complete chloroplast (cp) genome of V. schensianum based on Illumina paired-end sequencing for phylogenetic studies and the protection of genetic resources.
In this study, the fresh leaves of a single individual of V. schensianum were collected from Xianyang (Shaanxi, China; 108°04′E, 34°16′N) and Voucher herbarium specimens (SX10306) were deposited at the Herbarium of Guizhou Education University. Genomic DNA was extracted from the fresh leaves using the CTAB method (Doyle Citation1987). Total DNA was used for the shotgun library construction and the subsequent high-throughput sequencing on the Illumina HiSeq 2500 Sequencing System. In total, 2.7 G raw reads were obtained, quality-trimmed and used for the cp genome assembly using MITObim v1.8 (Hahn et al. Citation2013) with V. Japonicum (GenBank: MH036493.1) (Cho et al. Citation2018) as the initial reference. The genome was visualized and annotated in Geneious version 9.0.2 (Biomatters Ltd., Auckland, New Zealand). A neighbour-joining (NJ) tree was inferred using MEGA6.0 (Tamura et al. Citation2013) from alignments created by the MAFFT (Katoh and Standley Citation2013) using seven other complete chloroplast genomes previously reported in Adoxaceae. The annotated genomic sequence has been submitted to GenBank with the accession number MT003225.
The circular chloroplast genome of V. schensianum was 158,408 bp in size and comprises a pair of inverted repeat (IR) regions of 26,512 bp each, a large single-copy (LSC) region of 86,998 bp, and a small single-copy (SSC) region of 18,386 bp. The chloroplast genome contained 130 genes including 84 protein-coding genes, 37 tRNA genes, eight rRNA genes, and a pseudogene. In these genes, 18 genes contained one intron and two genes contained two introns. The majority of the gene species are single copy; however, 16 gene species in the IR regions are totally duplicated, including five protein-coding genes, seven tRNA genes, and four rRNA genes. Out of these 16 gene species, rps12 are partially located within the IR regions, whereas all the others completely within the IR regions. The overall GC content of V. schensianum chloroplast genome is 38.1%.
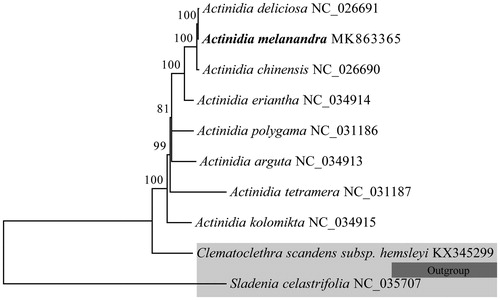
To investigate phylogeny of V. schensianum, phylogenetic analysis based on eight complete chloraplast genome sequences of Adoxaceae was performed using neighbour-joining (NJ) method with 1000 bootstrap replicates, of which Sinadoxa corydalifolia and Adoxa moschatellina were used as outgroup. As shown in the highly resolved NJ phylogenetic tree (), all the species of the genus Viburnum formed a monophyletic clade with a high resolution value and V. schensianum forms a clade with V. utile. The complete cp genome of V. schensianum would provide more valuable genomic information on metabolism, population genetics, molecular ecology, and evolution of Viburnum and related groups.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bibi Y, Nisa S, Waheed A, Zia M, Sarwar S, Ahmed S, Chaudhary MF. 2010. Evaluation of Viburnum foetens for anticancer and antibacterial potential and phytochemical analysis. Afr J Biotechnol. 9(34):5611–5615.
- Cho WB, Han EK, Choi HJ, Lee JH. 2018. The complete chloroplast genome sequence of Viburnum japonicum (Adoxaceae), an evergreen broad-leaved shrub. Mitochondrial DNA B Resour. 3(1):458–459.
- Clement WL, Arakaki M, Sweeney PW, Edwards EJ, Donoghue MJ. 2014. A chloroplast tree for viburnum (Adoxaceae) and its implications for phylogenetic classification and character evolution. Am J Bot. 101(6):1029–1049.
- Cometa MF, Mazzanti G, Tomassini L. 1998. Sedative and spasmolytic effects of Viburnum tinus L. and its major pure compounds. Phytother Res. 12(S1):S89–S591.
- Donoghue MJ, Baldwin BG, Li J, Winkworth RC. 2004. Viburnum phylogeny based on chloroplast trnK intron and nuclear ribosomal its DNA sequences. Syst Bot. 29(1):188–198.
- Donoghue MJ, Winkworth RC. 2005. Viburnum phylogeny based on combined molecular data: implications for taxonomy and biogeography. Am J Bot. 92(4):653–666.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol and Evol. 30(4):772–780.
- Lobstein A, Haan-Archipoff G, Englert J, Kuhry JG, Anton R. 1999. Chemotaxonomical investigation in the genus Viburnum. Phytochemistry (Oxford), 50(7):1175–1180.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wang GW. 2006. Resource of medicinal plants of Caprifo liaceae and its utilization in Ziwuling Forest District of Gansu Province. Lishizhen Med Mat Med Res. 17(9):1848–1850.