Abstract
The mitochondrial genome has proven to be a highly successful resource for the investigation of evolution and population genetics. Here, we present the complete mitochondrial genome of Fejervarya multistriata. The circle genome was found to be 17,759 bp in length, containing 13 protein-coding genes, 23 transfer RNA genes, 2 ribosomal RNA genes, and 1 noncoding control region that are conserved in most Dicroglossidae mitogenomes. The total base composition of the F. multistriata mitogenome is 28.04% A, 29.82% T, 26.99% C, and 15.15% G, which is typical for Amphibious animals’ mitochondrial genomes. Eight tRNAs are encoded on the light strand (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu and tRNA-Pro). Only one PCG is encoded on the light strand (ND6), whereas the other genes are located on the heavy strand. Phylogenetic analyses were performed on the concatenated dataset of 13 PCGs at nucleotide levels with maximum likelihood (ML) and Bayesian analysis (BI) methods. The results showed that F. multistriata and F. limnocharis integrated into a big branch and they have a close genetic relationship. This study could provide important molecular data for species identification and the phylogenetic relationship of Fejervarya and related species.
The paddy frog (Fejervarya multistriata) is a species widely distributed in the area from temperate to tropical Asia, belonging to the family Dicroglossidae (Frost Citation2018). There are many disputes about the classification of F. kawamurai and the closely related F. limnocharis and F. multistriata (Huang and Tu Citation2016). For example, F. kawamurai was a member of the F. limnocharis from western Honshu, Japan, and it is described as a new species of dicroglossid frog (Djong et al. Citation2011). The complete mitochondrial genome has proven to be a highly effective resource for the investigation of the genus Fejervarya species evolution and population genetics (Cheng et al. Citation2018). Thus, we sequenced the mitochondrial genome of F. multistriata to discuss the relationship within the genus Fejervarya phylogenetic analyses.
Specimens of F. multistriata were collected from Santai County, Mianyang City, Sichuan province, China, in August 2019 (105°7′49.89″E, 31°10′41.55″N), and immediately preserved in 95% ethanol at −80 °C until use. The clipped toe of the frog leg was taken and lysed by SDS/protease K and extracted by the phenol-chloride method (Ren et al. Citation2004) and deposited in the Key Laboratory for Molecular Biology and Biopharmaceutics, Mianyang Normal University (LC2019092206). We employed Long-and-Accurate PCR methods to amplify the whole mitogenomic region of F. multistriata with the self-designed and partial universal PCR primers for the mtDNAs of modern frogs (Liu et al. Citation2005; Kurabayashi and Sumida Citation2009). The sequence was submitted to the GenBank with the accession number MN733918.
The total length of the mitochondrial genome of F. multistriata is 17,759bp with a base composition of 28.04% A, 29.82% T, 26.99% C, and 15.15% G. The complete mitochondrial genome consists of 13 protein-coding genes, 2 rRNA genes, 23 tRNA genes, and 1 control region. The mitogenome of F. multistriata shows the typical gene content observed in vertebrate mitogenomes (Maurino and Viscusi Citation2009; Zhou et al. Citation2009; Li et al. Citation2014). Eight tRNAs are encoded on the light strand (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu and tRNA-Pro). Only one PCG is encoded on the light strand (ND6), whereas the other genes are located on the heavy strand. Most protein-coding genes begin with ATG as the start codon, except for ND2 gene with ATC, COI gene with ATA, ND3 gene with GTG, and ND3 gene with GTG. The ND6 gene is terminated with AGG as the stop codon, whereas ND1, ND2, ND5, COI, COII, ATP6, and COIII genes end with an incomplete stop codon (T––) and the other protein-coding genes end with TAA.
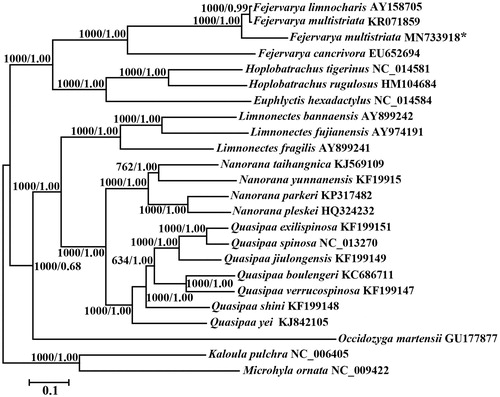
To elucidate phylogenetic relationships of F. multistriata with the other related species, phylogenetic trees were reconstructed using BI and ML methods based on the nucleotide dataset (Alam et al. Citation2010; Yu et al. Citation2012). The best-fit GTR + I + G model was selected in jModelTest 0.1 (Darriba et al. Citation2012), and yielded identical phylogenetic trees by high node-supporting values, including that 22 reported dicroglossids frogs (). In the phylogenetic tree, F. multistriata clustered with F. limnocharis into a branch and they have a close genetic relationship. The monophyly of Fejervarya, Quasipaa, Limnonectes and Nanorana was well supported as also reported in other recent studies (Zhang et al. Citation2009; Cai et al. Citation2018). In this study, we present the complete mitochondrial genome sequence of F. multistriata, which would contribute to further phylogenetic analysis of this species. More mitochondrial genomic data of undetermined taxa and further analysis are required to reveal the phylogeny and evolution of Dicroglossidae.
Figure 1. Phylogenetic tree of the relationships among 22 dicroglossids frogs and two species of Microhylidae as outgroups (Microhyla pulchra and Kaloula pulchra) based on the nucleotide dataset of the 13 mitochondrial protein-coding genes. Branch lengths and topology are from the BI analysis. Numbers above branches specify posterior probabilities from Bayesian inference (BI) and bootstrap percentages from maximum likelihood (ML, 1000 replications) analyses. Tree topologies produced by Bayesian inferences (BI) and maximum likelihood (ML) analyses were equivalent. bootstrap support values for ML analyses and Bayesian posterior probability are shown orderly on the nodes. The asterisks indicate new sequences generated in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alam MS, Kurabayashi A, Hayashi Y, Sano N, Khan MMR, Fujii T, Sumida M. 2010. Complete mitochondrial genomes and novel gene rearrangements in two dicroglossid frogs, Hoplobatrachus tigerinus and Euphlyctis hexadactylus, from Bangladesh. Genes Genet Syst. 85(3):219–232.
- Cai Y-T, Ma L, Xu C-J, Li P, Zhang J-Y, Storey K B, Yu D-N. 2018. The complete mitochondrial genome of the hybrid of Hoplobatrachus chinensis (♀)× H. rugulosus (♂) and its phylogeny. Mitochondrial DNA Part B. 3(1):344–345.
- Cheng JX, Cai YT, Zheng YJ, Zhang JY, Storey KB, Bao YX, Yu DN. 2018. The complete mitochondrial genome of Fejervarya kawamurai (Anura: Dicroglossidae) and its phylogeny. Mitochondrial DNA Part B. 3(2):551–553.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Djong HT, Matsui M, Kuramoto M, Nishioka M, Sumida M. 2011. A new species of the Fejervarya limnocharis complex from Japan (Anura, Dicroglossidae). Zool Sci. 28(12):922–929.
- Frost DR. 2018. Amphibian species of the world: an online reference. New York: American Museum of Natural History. [accessed 2018, Mar 1]. http://research.amnh.org/herpetology/amphibia/index.html.
- Huang ZH, Tu FY. 2016. Mitogenome of Fejervarya multistriata: a novel gene arrangement and its evolutionary implications. Genet Mol Res. 15:15038302.
- Kurabayashi A, Sumida M. 2009. PCR primers for the neobatrachian mitochondrial geneome. Curr Herpet. 2:1–11.
- Li E, Li X, Wu X, Feng G, Zhang M, Shi H, Wang L, Jiang J. 2014. Complete nucleotide sequence and gene rearrangement of the mitochondrial genome of Occidozyga martensii. J Genet. 93(3):631–641.
- Liu ZQ, Wang YQ, Su B. 2005. The mitochondrial genome organization of the rice frog, Fejervarya limnocharis (Amphibia: Anura): a new gene order in the vertebrate mtDNA. Gene. 346:145–151.
- Maurino A, Viscusi G. 2009. Complete nucleotide sequence and gene arrangement of the mitochondrial genome of the crab-eating frog Fejervarya cancrivora, and evolutionary implications. Gene. 441:148–155.
- Ren ZM, Ma EB, Guo YP, Zhong Y. 2004. A molecular phylogeny of Oxya (Orthoptera: Acridoidea) in China inferred from partial cytochrome b gene sequences. Mol Phylogenet Evol. 33(2):516–521.
- Yu D, Zhang J, Zheng R, Shao C. 2012. The complete mitochondrial genome of Hoplobatrachus rugulosus (Anura: Dicroglossidae). Mitochondrial DNA. 23(5):336–337.
- Zhang JF, Nie LW, Wang Y, Hu LL. 2009. The complete mitochondrial genome of the large-headed frog, Limnonectes bannaensis (Amphibia: Anura), and a novel gene organization in the vertebrate mtDNA. Gene. 442(1-2):119–127.
- Zhou Y, Zhang JY, Zheng RQ, Yu BG, Yang G. 2009. Complete nucleotide sequence and gene organization of the mitochondrial genome of Paa spinosa (Anura: Ranoidae). Gene. 447(2):86–96.
