Abstract
The genus Cucumis contains 52 species, including two economically significant crops, cucumber, and melon, as well as some important landraces. Cucumis melo L. ‘Shengkaihua’ is a widely cultivated landraces in China. It has high sugar content, early maturity, and strong genetic growth potential. Cucumis melo L. ‘Shengkaihua’ provides a valuable gene pool for crop improvement of Cucumis crops. In this study, the complete chloroplast (cp) genome sequence of C. melo L. ‘Shengkaihua’ was determined using next-generation sequencing. The entire cp genome was determined to be 156,017 bp in length. It contained large single-copy (LSC) and small single-copy (SSC) regions of 86,335 and 18,088 bp, respectively, which were separated by a pair of 25,797 bp inverted repeat (IR) regions. The genome contained 133 genes, including 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content of the genome is 36.9%. A phylogenetic tree reconstructed by 24 chloroplast genomes reveals that C. melo L. ‘Shengkaihua’ is most related to Cucumis melo var. inodorus.
The genus Cucumis contains 52 species, including two economically significant crops, cucumber, and melon, as well as some important landraces. Cucumis melo L. ‘Shengkaihua’ is a widely cultivated landraces in China (Luan et al. Citation2008). It has high sugar content, early maturity, and strong genetic growth potential. Cucumis melo L. ‘Shengkaihua’ provides a valuable gene pool for crop improvement of Cucumis crops. However, genetic relationships between C. melo L. ‘Shengkaihua’ and other melon species on genomes level have not been studied. So, it is necessary to develop genomic resources for C. melo L. ‘Shengkaihua’ to provide basic intragenic information for the further study on phylogeny and breeding for genus Cucumis.
The total genomic DNA was extracted from the fresh leaves of C. melo L. ‘Shengkaihua’ (Gaolan County, Gansu, China, 36.6 N, 103.38E) using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). The voucher specimen was deposited at Gansu Academy of Agricultural Sciences (2016SKH). The whole genome sequencing was conducted by Genepioneer Biotechnologies Inc. (Nanjing, China) on the Illumina Hiseq 4000 Sequencing System (Illumina, Hayward, CA). The filtered sequences were assembled using the program SPAdes assembler 3.10.0 (Bankevich et al. Citation2012). Annotation was performed using the DOGMA (Wyman et al. Citation2004). All the tRNA sequences were confirmed using the web-based online tool, tRNAScan-SE (Schattner et al. Citation2005) with default settings to corroborate tRNA.
The plastome of C. melo L. ‘Shengkaihua’ was determined to comprise double-stranded, circular DNA of 156,017 bp containing two inverted repeat (IR) regions of 25,797 bp each, separated by large single-copy (LSC) and small single-copy (SSC) regions of 86,335 and 18,088 bp, respectively (NCBI acc. no. MN990709). The genome contained 133 genes, including 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The seven protein-coding genes, seven tRNA genes, and four rRNA genes were duplicated in IR region. Seventeen genes contained two exons and four genes (clpP and ycf3 and two rps12) contained three exons. The overall GC content of C. melo L. ‘Shengkaihua’ cp genome is 36.9% and the corresponding values in LSC, SSC, and IR regions are 34.7, 30.9, and 42.8%, respectively.
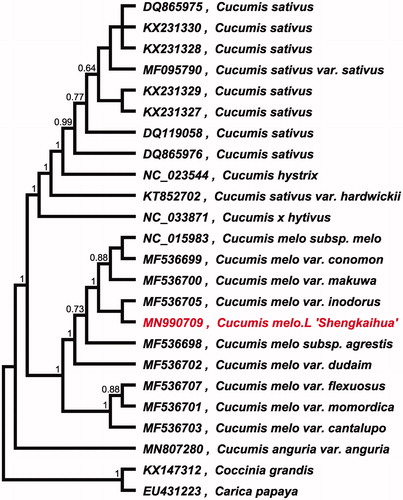
To investigate its taxonomic status, whole chloroplast genomes from 22 Cucumis plants, and two outgroup plants (Coccinia grandis and Carica papaya) were aligned using MAFFT version 7 (Katoh and Standley Citation2013). A maximum-likelihood (ML) was reconstructed based on FastTree version 2.1.10 (Price et al. Citation2010). The ML phylogenetic tree shows that C. melo L. ‘Shengkaihua’ is most related to Cucumis melo var. inodorus ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Luan F, Delannay I, Staub JE. 2008. Chinese melon (Cucumis melo L.) diversity analyses provide strategies for germplasm curation, genetic improvement, and evidentiary support of domestication patterns. Euphytica. 164(2):445–461.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 5(3):e9490.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.